This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Metabolism meets signaling to fine-tune cell growth
Cells use a variety of metabolic pathways to synthesize the building blocks for growth and proliferation. To ensure balanced growth, these biosynthetic processes must be tightly coordinated. Researchers from the Max Planck Institute for Biology of Ageing, together with a team of national and international collaborators, have now identified a molecular machinery that senses the ability of a cell to make lipids to then block or activate all other biosynthetic processes, such as protein synthesis, accordingly.
Cells accumulate mass and grow by using nutrients and energy to build membranes, proteins, nucleic acids, and other macromolecular structures, through the coordinated action of various metabolic pathways. Therefore, nutrient sensing mechanisms ensure that cells only grow when all conditions are optimal.
The most important nutrient sensor in cells is a protein complex called mTORC1. When amino acids, the building blocks of proteins, are abundant, mTORC1 is active and promotes protein synthesis, thus linking amino acid availability to the respective cellular function. However, how the ability of cells to make lipids is sensed and whether mTORC1 plays a role in this process has been very poorly understood.
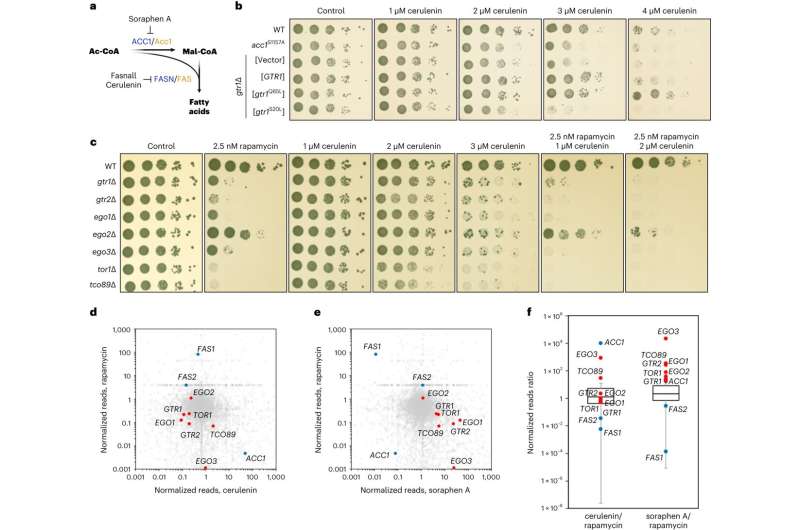
In fatty acid biosynthesis, acetyl-CoA, a metabolite originating mainly from the breakdown of glucose (another important nutrient), is first converted to malonyl-CoA by an enzyme called ACC1, which is then converted to fatty acids by an enzyme called FASN (fatty acid synthase).
The researchers showed that accumulation of malonyl-CoA, which can occur when FASN levels or activity are low, leads to inhibition of mTORC1 activity and downregulation of other biosynthetic functions that are regulated by mTORC1, such as protein synthesis. Surprisingly, they discovered that malonyl-CoA binds directly to the catalytic pocket of mTOR, competing away ATP, another metabolite that is necessary for mTOR to be active and phosphorylate its substrates.
"These findings highlight malonyl-CoA as the first described endogenous metabolite that acts as an ATP-competitive inhibitor for a signaling kinase in mammalian cells," says Constantinos Demetriades, research group leader at the Max Planck Institute for Biology of Ageing and head of the study.
"It is noteworthy that this phenomenon is evolutionarily conserved from yeast to human cells, thus it represents an ancient mechanism via which lipid metabolism talks to the core regulator of all other metabolic processes, mTORC1."
Linking fatty acid biosynthesis to cell growth through mTORC1
At the molecular level, the researchers found that mTORC1 forms physical interactions with ACC1 and FASN, allowing it to directly sense the levels of malonyl-CoA at the site where the metabolite is produced. In this way, the direct binding of malonyl-CoA to mTORC1 switches off all other cellular processes that are controlled downstream of this complex.
As a result, under conditions that a cell cannot produce enough fatty acids, mTORC1 inhibition would also block synthesis of proteins, other anabolic processes and, eventually, cell growth, until conditions are optimal again.
As pharmacological inhibitors of FASN, like those used in this study, are used as anti-cancer drugs in the clinic, these discoveries may also be relevant for cancer treatment in the future. Based on the data described here, FASN inhibitors likely have a two-fold effect in cancer cells: not only do they block fatty acid synthesis, which is required for membrane formation and energy production, but they also inhibit mTORC1 activity, which is dysregulated in most cancers.
The paper is published in the journal Nature Cell Biology.
More information: Raffaele Nicastro et al, Malonyl-CoA is a conserved endogenous ATP-competitive mTORC1 inhibitor, Nature Cell Biology (2023). DOI: 10.1038/s41556-023-01198-6