This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Scientists find evidence of unintended impacts from anti-cancer drugs
With around 90% of drugs failing to make it to market, the potential for improving efficiency within the drug development industry is clear. Drugs designed to combat cancers suffer similar rates of failure for many reasons. Now, researchers have revealed one reason why certain anti-cancer compounds can cause unexpected side effects. This research could help guide an understanding of why some drugs show more promise than others, providing a new tool that can be used to identify those drugs and drug candidates.
One of the most essential and energy-consuming cellular processes is ribosome biogenesis, the formation of the cellular machines that manufacture all proteins. For cancer cells, this process is paramount. A recent study published in eLife from the Stowers Institute for Medical Research screened over 1,000 existing anti-cancer drugs to assess how they impact the structure and function of the nucleolus, the ubiquitous cellular organelle where ribosomes are made.
"All cells must make proteins to function, so they have to make ribosomes, which are also protein complexes themselves," said lead author Tamara Potapova, Ph.D., a research specialist in the lab of Investigator Jennifer Gerton, Ph.D. "In cancer cells, ribosome production must be in overdrive to compensate for high proliferation rates requiring even more proteins."
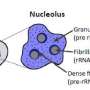
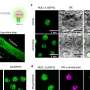
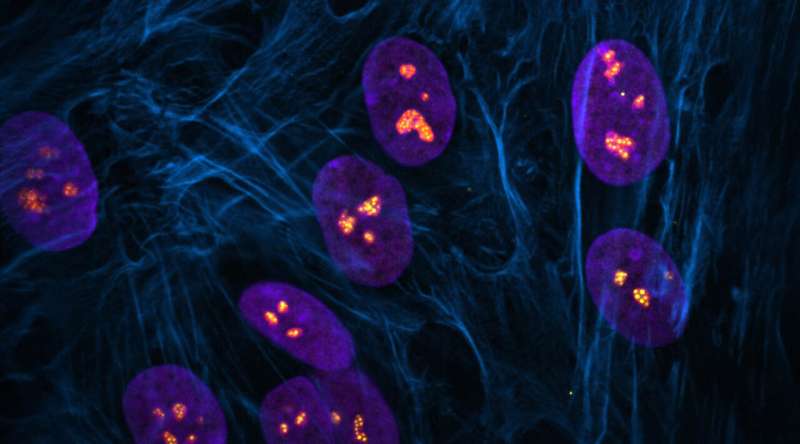
The nucleolus is a special part of the cell nucleus that houses ribosomal DNA, and where ribosomal RNA production and ribosome assembly largely takes place. Nucleoli can vary greatly in appearance, serving as visual indicators of the overall health of this process. Thus, the team found a way to capitalize on this variation and asked how chemotherapy drugs impact the nucleolus, causing nucleolar stress.
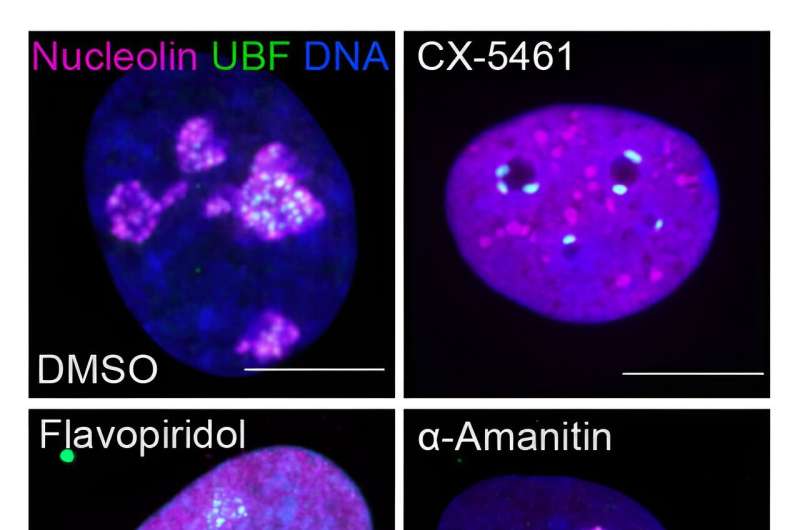
"In this study, we not only evaluated how anti-cancer drugs alter the appearance of nucleoli, but also identified categories of drugs that cause distinct nucleolar shapes," said Gerton. "This enabled us to create a classification system for nucleoli based on their appearance that is a resource other researchers can use."
Because cancer's hallmark is unchecked proliferation, most existing chemotherapeutic agents are designed to slow this down. "The logic was to see whether these drugs, intentionally or unintentionally, are affecting ribosome biogenesis and to what degree," said Potapova. "Hitting ribosome biogenesis could be a double-edged sword—it would impair the viability of cancer cells while simultaneously altering protein production in normal cells."
Different drugs impact different pathways involved in cancer growth. Those that influence ribosome production can induce distinct states of nucleolar stress that manifest in easily seen morphological changes. However, nucleolar stress can be difficult to measure.
"This was one of the issues that impeded this field," said Potapova. "Cells can have different numbers of nucleoli with different sizes and shapes, and it has been challenging to find a single parameter that can fully describe a 'normal' nucleolus. Developing this tool, that we termed 'nucleolar normality score,' allowed us to measure nucleolar stress precisely, and it can be used by other labs to measure nucleolar stress in their experimental models."
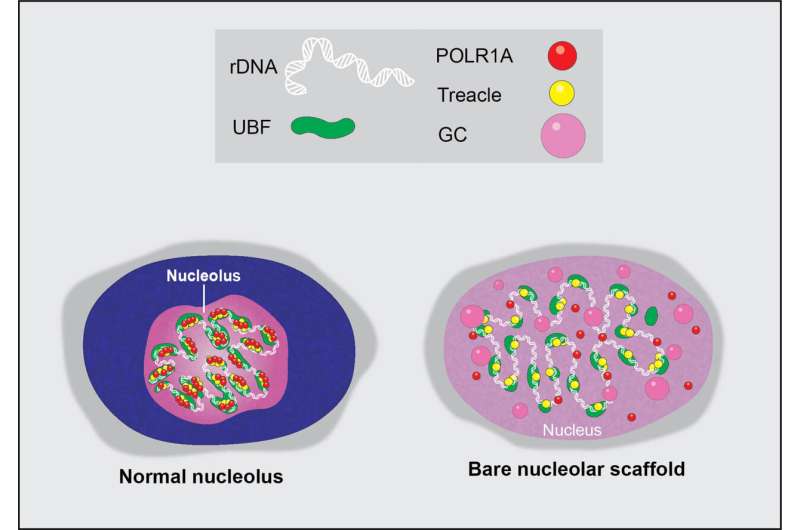
Through the comprehensive screening of anti-cancer compounds on nucleolar stress, the team identified one class of enzymes in particular, cyclin-dependent kinases, whose inhibition destroys the nucleolus almost completely. Many of these inhibitors failed in clinical trials, and their detrimental impact on the nucleolus was not fully appreciated previously.
Drugs often fail in clinical trials due to excessive and unintended toxicity that can be caused by their off-target effects. This means that a molecule designed to target one pathway may also be impacting a different pathway or inhibiting an enzyme required for cellular function. In this study, the team found an effect on an entire organelle.
"I hope at a minimum this study increases awareness that some anti-cancer drugs can cause unintended disruption of the nucleolus, which can be very prominent," said Potapova. "This possibility should be considered during new drug development."
More information: Tamara A. Potapova et al, Distinct states of nucleolar stress induced by anti-cancer drugs, eLife (2023). DOI: 10.7554/eLife.88799.1