This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
Researchers uncover aberrant natural killer cell subtype
A team led by Prof. Tian Zhigang and Prof. Peng Hui from the University of Science and Technology of China (USTC) of the Chinese Academy of Sciences (CAS), in collaboration with Prof. Zhang Zemin from Peking University, have depicted the heterogeneity of natural killer (NK) cells across different cancer types and tissues, and discovered a subset of NK cells with aberrant anti-tumor functions that thrive specifically within the tumor microenvironment. The study was published in Cell.
The NK cell, named after its ability to directly kill cancer cells, has emerged as a formidable contender in immunotherapy, showcasing exceptional efficacy in blood cancer treatments. However, the heterogeneity of NK cells, varying in phenotype and function within distinct tissue microenvironments, has posed challenges in its application in solid tumor therapy.
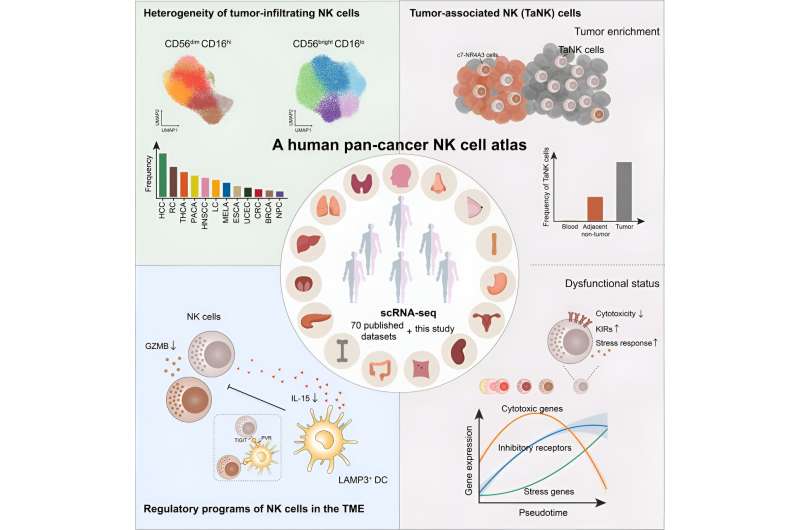
Over the years, Prof. Tian and Prof. Peng's team have been studying the heterogeneity of NK cells across different tissues. In this study, the researchers collected an extensive dataset of single-cell transcriptomes, encompassing 24 cancer types and including a total of 1,223 samples from 716 patients and 47 healthy individuals.
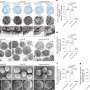
They identified five distinct subtypes of CD56brightCD16lo NK cells and nine subtypes of CD56dimCD16hi NK cells at a comprehensive pan-cancer level for the first time. These subtypes were meticulously characterized for their phenotypic and functional diversity.
Through the integration of this extensive dataset, the researchers observed a preference of NK cell subtype composition across different cancer types. Notably, the distribution of NK cell subtypes within tumors, neighboring tissues and peripheral blood displayed significant disparities.
Leveraging advanced bioinformatics techniques, the researchers pinpointed the gene RGS1 as being highly expressed in non-blood NK cells. At transcriptional level, RGS1 demonstrated remarkable specificity and sensitivity, compared to conventional tissue residency markers.
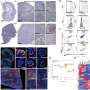
By probing the tumor microenvironment, the researchers found that a group of DNAJB1+CD56dimCD16hi NK cells were highly enriched in tumor issues. Analysis of this group revealed a dysfunctional phenotype with reduced cytotoxicity, increased inhibitory receptors, and heightened stress-related protein levels.
This subtype, named "Tumor-associated NK cells" (TaNK cells), defied the conventional understanding that higher NK cell abundance is beneficial to tumor patients. Instead, TaNK cells exhibited a strong association with adverse prognoses in various cancer types and a significant resistance to immunotherapy.
Additionally, the researchers discovered that LAMP3+ dendritic cells (DCs) are critical regulators of NK cell functionality. Spatial distribution data analysis showed that NK cells in close proximity to LAMP3+ DCs exhibited diminished cytotoxic activity. This observation hinted at the potential of LAMP3+ DCs to exert abnormal regulatory effects on NK cell function in the tumor microenvironment.
More information: Fei Tang et al, A pan-cancer single-cell panorama of human natural killer cells, Cell (2023). DOI: 10.1016/j.cell.2023.07.034