This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Histone acetylation, transcription factor dynamics contribute to gene expression in brain development
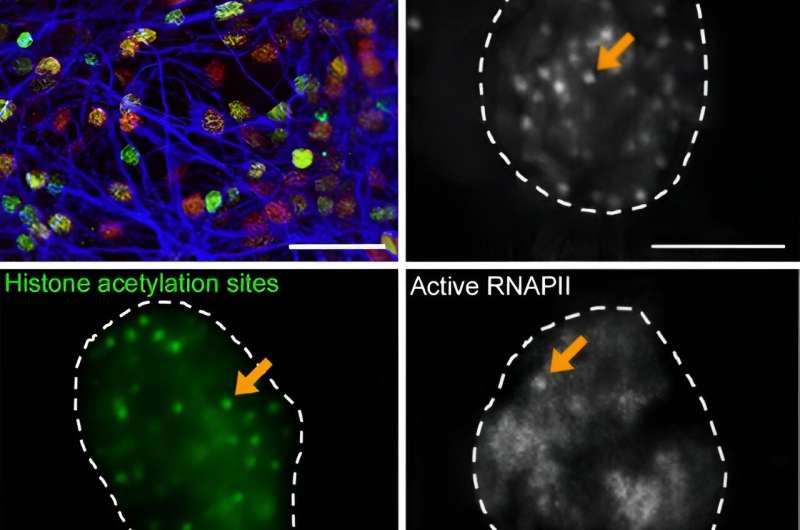
No man is an island, and no neuron is either: complex, intricate connections between and among brain cells are needed for the brain to do its job correctly. Now, researchers from Japan have uncovered a fundamental mechanism that lets neurons know when and how to form these connections.
In the study "Repetitive CREB-DNA interactions at gene loci predetermined by CBP induce activity-dependent gene expression in human cortical neurons," recently published in Cell Reports, researchers from Osaka University and Shenzhen Bay Laboratory have revealed the behavior of proteins linked to learning and memory in human brain cells.
In the developing brain, neuronal activity stimulates the expression of genes that are needed for brain cells to grow and form connections. If this process is disrupted, developmental and psychiatric disorders can develop.
"The transcription factor cAMP response element binding protein (CREB) is required for the brain to learn and form memories," says lead author of the study Yuri Atsumi. "While CREB and CREB binding protein (CBP) are known to interact to stimulate gene expression in the developing brain, it is unclear how neuronal activity affects this interaction."
To explore this, the researchers used a sophisticated microscopy technique called single-molecule imaging to track the movements of CREB and CBP in human brain cells.
"The results were very clear," explains co-senior author Noriyuki Sugo. "We found that neuronal activity markedly increased repetitive, long-term interactions between CREB and DNA and between CREB and CBP."
These interactions attracted RNA polymerase II, which activates gene expression. Furthermore, the researchers discovered that the sites where CREB interacted with DNA to promote gene expression require prior acetylation mediated by CBP.
"These findings not only elucidate the mechanism underlying learning and memory in developing brain, but also offer new insight into how psychiatric disorders may develop," says co-senior author Nobuhiko Yamamoto.
Given that CBP is a gene that can cause Rubinstein–Taybi syndrome, in which patients show mental retardation and intellectual disability, inactivation of the CBP protein could help explain how this disease develops. Exploring CREB-CBP interactions further in 3D brain-like models could yield additional insight into the effects of neuronal activity observed in this study.
More information: Yuri Atsumi et al, Repetitive CREB-DNA interactions at gene loci predetermined by CBP induce activity-dependent gene expression in human cortical neurons, Cell Reports (2023). DOI: 10.1016/j.celrep.2023.113576