This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Colorectal cancer gene implicated in broad range of solid tumors
A gene associated with colorectal cancer appears to also play a role in the development of other solid tumors, according to a study of over 350,000 patient biopsy samples conducted by researchers at the Johns Hopkins Kimmel Cancer Center, the Johns Hopkins Bloomberg School of Public Health and Foundation Medicine.
Since the early 2000s, scientists have known that inheriting two mutated copies of the gene MUTYH leads to a 93-fold increased risk of colorectal cancer and is a major cause of that cancer in individuals younger than 55. The new study, published in JCO Precision Oncology, is the largest analysis to date to investigate whether a single mutated copy of MUTYH also affects one's risk of developing cancer.
"We know two missing copies of MUTYH greatly increases the risk of colon cancer, and now it appears that having only one missing copy may lead to a small increased risk of other cancer types," says the study's lead author, Channing Paller, M.D., director of prostate cancer clinical research and an associate professor of oncology at the Johns Hopkins University School of Medicine.
She co-led the work with Emmanuel Antonarakis, M.D., associate director of translational research at the Masonic Cancer Center, and Clark Endowed Professor of Medicine at the University of Minnesota Medical School. He was at Johns Hopkins at the time the research was conducted.
The gene MUTYH encodes a critical enzyme in the base excision repair (BER) pathway, which fixes DNA damage in human cells. When the BER pathway isn't working, routine DNA damage is not repaired, leading to additional DNA mutations or cell death.
Since 2021, Paller has co-led PROMISE, a genetic registry of patients with inherited mutations in prostate cancer. When one of her patients asked whether his MUTYH mutation, for which he had one defective copy rather than two, affected his aggressive prostate cancer, there was not enough data on MUTYH variants to answer the question, says Paller. Past studies reached conflicting results about whether a single, heterozygous mutation of MUTYH might predispose a person to cancer.
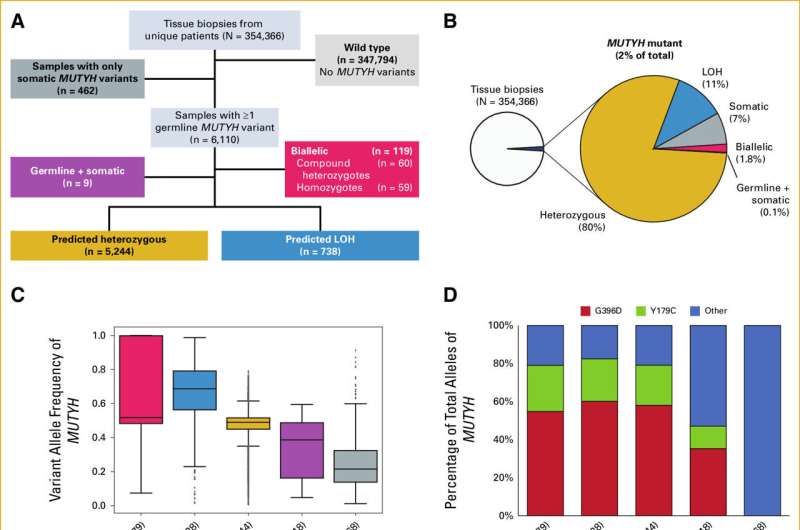
In pursuit of an answer, Paller reached out to Foundation Medicine, a Massachusetts-based genomic profiling company that maintains one of the world's largest cancer genomic databases. With researchers at Foundation Medicine; Alexandra Maertens, Ph.D., of the Center for Alternatives to Animal Testing at the Bloomberg School of Public Health; and others, the team applied an advanced algorithm to analyze the genetic data of 354,366 solid tumor biopsies stored in the Foundation database.
Within that population of tumor samples, 5,991 had one working version and one mutated version of MUTYH. Of those, 738 (about 12%) had lost their working copy of the gene, leaving them with just the mutated copy.
Those with a single, mutated copy of MUTYH showed a genetic signature, like a fingerprint, of additional genetic mutations and a defective BER pathway. Individuals with that genetic signature had a modest increase in susceptibility to a subset of solid tumors, including adrenal gland cancers and pancreatic islet cell tumors. However, they did not have an increased risk for breast or prostate cancer, resolving the original patient's question.
The results suggest that MUTYH variants might be involved in a broader range of cancers than previously known, Paller says.
"The next question is whether this finding has therapeutic implications," she says. "Can we target the BER pathway for possible drug sensitivities?" If so, doctors might be able to add a new therapeutic approach to their arsenal of tools against solid cancers.
More information: Channing J. Paller et al, Pan-Cancer Interrogation of MUTYH Variants Reveals Biallelic Inactivation and Defective Base Excision Repair Across a Spectrum of Solid Tumors, JCO Precision Oncology (2024). DOI: 10.1200/PO.23.00251