This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Discovery sets stage for vaccine against gastric cancer, ulcers
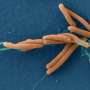
H. pylori is one of the most common disease-causing bacteria. More than half of the world's population have the bacteria in their body; and while in Canada overall prevalence of H. pylori is between 20% and 30%, some groups—including Indigenous communities—have higher rates.
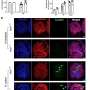
Using the Canadian Light Source at the University of Saskatchewan (USask) researchers from Quebec's National Institute of Scientific Research (INRS) have for the first time solved the structure of the protein that plays a key role in helping H. pylori stick to the lining of our stomach. Their research paves the way for developing a vaccine against the infection.
It is H. pylori's ability to bind to the inside of the stomach that helps it survive and cause health problems. The pathogen is responsible for nearly all gastric cancers and peptic ulcers. Around one in 10 people who carry the common pathogen will develop an ulcer; almost 3% will get stomach cancer.
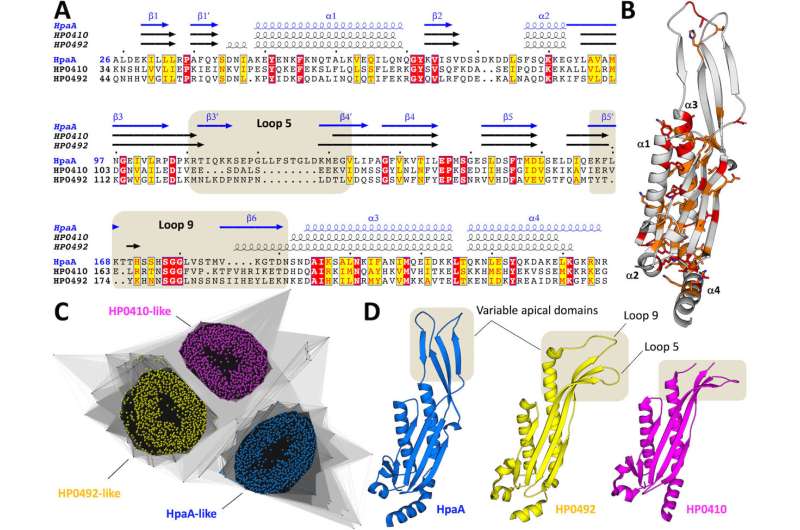
Professor Charles Calmettes, a biochemist at INRS, says that being able to see the structure of the protein HpaA helps scientists better understand H. pylori's "stickiness" and why our body reacts by causing certain immune cells to cause inflammation. His team's findings were published in the journal mBio.
"Investigating this bacterium is a way to help find new vaccine or drug targets, because there is some concern about antibiotic resistance," says Calmettes. "For the future, we will need to have some more tools to treat this infection."
Calmettes says the CLS's CMCF beamline was key in understanding how the protein binds a receptor on to a human cell. "Most of our studies rely on solving the atomic structure of bacterial virulence factors. And for this we need the synchrotron light," says Calmettes. "Without the CLS we are not able to solve the structure of HpaA."
More information: Cyrielle Martini et al, Unraveling the crystal structure of the HpaA adhesin: insights into cell adhesion function and epitope localization of a Helicobacter pylori vaccine candidate, mBio (2024). DOI: 10.1128/mbio.02952-23