This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
New protein target identified for early Alzheimer's treatment
A class of proteins that regulates cell repair and enhances cell growth-signaling systems could be a promising new target for the treatment of Alzheimer's and other neurodegenerative diseases, according to a new study led by researchers at Penn State. They found that disrupting necessary sugar modifications of these proteins promotes cell repair and reverses cellular abnormalities that occur in neurodegenerative diseases.
The study appeared July 2 in the journal iScience, and the researchers have a patent related to this work.
"Strategies to treat Alzheimer's disease to date have largely focused on pathological changes prominent in the late stages of the disease," said Scott Selleck, professor of biochemistry and molecular biology in the Penn State Eberly College of Science and leader of the research team.
"Although recently [U.S. Food and Drug Administration]-approved drugs have shown the ability to modestly slow the disease by targeting one of these changes, amyloid accumulation, drugs that affect the earliest cellular deficits might provide important tools to stop or reverse the disease process.
"We are interested in understanding the earliest cellular changes that are found not only in Alzheimer's, but shared across other neurodegenerative diseases, including Parkinson's and amyotrophic lateral sclerosis (ALS)."
Roughly 6.9 million Americans over the age of 65 are estimated to be living with Alzheimer's disease, according to the Alzheimer's Association. Despite its widespread impact, there is no agreed upon biological cause or mechanism for the disease.
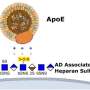
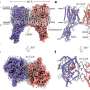
Cell-signaling molecules called heparan sulfate–modified proteins have been implicated in the development of Alzheimer's, but their specific role has remained unclear, Selleck said. In this study, the research team first performed a series of analyses in human cell lines and mouse brain cells that express aspects of Alzheimer's, showing that these proteins regulate cellular processes known to be affected in several neurodegenerative diseases.
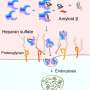
Heparan sulfate–modified proteins are found both on the surface of animal cells and in the matrix between cells. This class of proteins are named for a sugar polymer that bears many sulfate groups, called heparan sulfate.
Heparan sulfate chains are attached to specific proteins, and this modification allows these proteins to assemble signaling complexes that affect cell growth and influence how the cell interacts with its environment. These signaling pathways also regulate autophagy, a process of cell repair that clears out damaged or dysfunctional components in the cell.
"In the early stages of several neurodegenerative diseases, autophagy is compromised, which means cells have a reduced repair capacity," Selleck said.
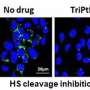
"In this study, we determined that heparan sulfate-modified proteins suppress autophagy-dependent cell repair. What's more, we show that by compromising the structure and function of the sugar modifications of these proteins, the levels of autophagy increase so cells can take care of damage."
The researchers found that, in human and mouse cells, reducing the function of heparan sulfate-modified proteins also rescued other pathologies that arise early in neurodegenerative diseases, improving the function of mitochondria—which are responsible for energy production in the cell—and reducing build-up of lipids, or fatty compounds, inside cells.
The researchers then evaluated the role of heparan sulfate-modified proteins in an animal model of Alzheimer's, a fruit fly with deficits in a presenilin protein. Presenilin mutations cause early onset disease in humans and likewise in fruit flies; defective presenilin causes cell death and brain degeneration.
In flies with deficits in presenilin, reducing the function of heparan sulfate chains suppressed the death of neurons and corrected other cell defects as well. These results are directly relevant to recent human genetics research, the researchers said.
"Individuals with mutations in a presenilin gene, PSEN1, develop Alzheimer's in their mid-40s. But if they also inherit a rare genetic change in a specific protein called APOE, the disease is delayed, sometimes by decades," Selleck said, explaining that APOE plays an important role in lipid transport and binds to heparan sulfate.
"This change in APOE—which has been in the news lately—greatly reduces APOE binding to heparan sulfate. Our work builds on and extends these findings, directly implicating heparan sulfate in Alzheimer's pathology involving both PSEN1 and APOE. Targeting the enzymes that make heparan sulfate could provide a means of blocking neurodegeneration in humans."
Collectively, these results show that disrupting the structure of heparan sulfate modifications, blocks or reverses early cellular problems in these models of Alzheimer's.
"We save the animal from neuron cell loss, mitochondrial defects and rescue behavior deficits that serve as a measure of nervous system function," Selleck said. "These findings suggest a promising target for future treatments that could rescue the earliest abnormalities that occur in many neurodegenerative diseases."
The researchers also explored how gene expression changed when they eliminated the capacity of human cells to make heparan sulfate chains. They found that expression levels of more than 50% of approximately 70 genes known to be associated with late-onset Alzheimer's disease were modulated, including APOE, suggesting a link between heparan sulfate-modified proteins and the more common and late onset forms of Alzheimer's disease.
"There is a critical need to focus on cellular changes that occur at the earliest times in disease progression and develop treatments that block or reverse them," Selleck said.
"We demonstrate that reduced autophagy, mitochondrial defects and lipid build-up—all common changes in neurodegenerative disease—can be blocked by altering one class of proteins, those with heparan sulfate modifications. We think these molecules are promising targets for drug development."
The researchers suspect that disrupting this pathway to promote cell repair systems could be important for a wide variety of other diseases where autophagy defects occur.
"The applications of manipulating this pathway may be broadly useful across a number of human medical conditions," Selleck said.
More information: Altering heparan sulfate suppresses cell abnormalities and neuron loss in Drosophila presenilin model of Alzheimer's, iScience (2024). DOI: 10.1016/j.isci.2024.110256. www.cell.com/iscience/fulltext … 2589-0042(24)01481-0