This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Machine learning may lead to better flu vaccines
A team led by scientists at UGA's Odum School of Ecology has developed an algorithm that can accurately predict how a seasonal flu virus is expected to evolve. Such information may allow seasonal flu vaccines to be updated more quickly, leading to reduced infections and deaths.
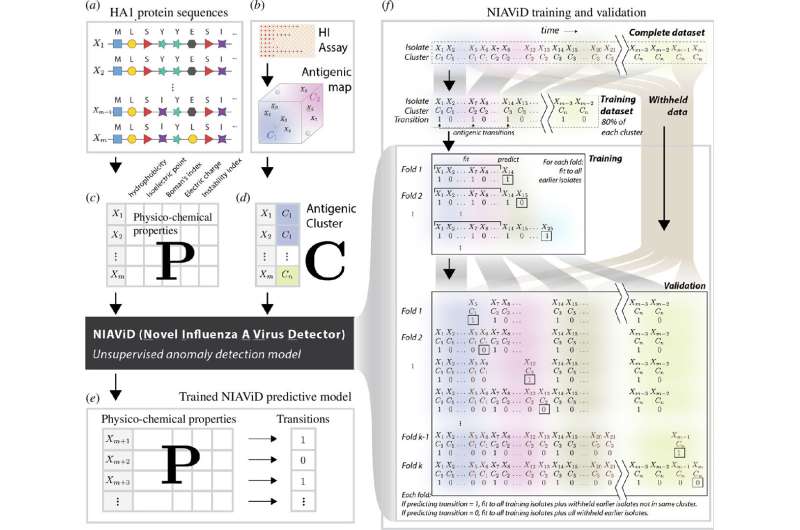
Published in the journal Proceedings of the Royal Society B: Biological Sciences, the research describes a machine-learning tool named Novel Influenza Virus A Detector (NIAViD) that predicts changes in the seasonal flu virus with nearly 73% accuracy. Alpha Forna, a postdoctoral research associate in the Odum School, led the development of NIAViD, which uses flu virus sequence data to understand how emerging viruses are expected to bypass existing immunity in a population.
Each year, seasonal flu viruses globally infect 1 billion people, and the majority of the estimated 500,000 deaths are young children in developing countries. In the United States, millions of Americans are infected annually, and thousands die. The projected annualized cost of seasonal infections is around $90 billion in just the United States.
One of the challenges in preventing seasonal flu infections is predicting how a flu virus is expected to change, according to Forna.
"Each flu virus, which we call influenza, has a hemagglutinin protein that initiates an infection when it attaches to cells. During an infection, influenza viruses begin to replicate, and as this happens, small genetic changes to the hemagglutinin protein occur, which we call antigenic drift," he said.
"Some of these changes may make an influenza virus unrecognizable to our immune system. This means that although someone may have had a previous influenza infection or received a vaccine, they might still be infected by an influenza virus that has a slightly evolved hemagglutinin protein."
Machine learning, a method of programming computers to learn on their own, is an increasingly important tool in the field of infectious disease modeling research. Forna spent two years building the NIAViD system, accessing and making sense of large datasets and training models for accurate predictions. He and his colleagues developed a specific set of algorithms that were designed to analyze amino acid sequences in a region of the hemagglutinin protein's gene and several associated properties that quantify traits, for example, a virus's electrostatic charge.
After the model was trained on a subset of data, its accuracy rate was nearly 73%, equaling or exceeding the performance of other models. By focusing on key properties, NIAViD precisely identifies antigenic changes, aiding in the timely update of flu vaccines.
NIAViD was developed around an influenza virus that first emerged during the 1968 pandemic that started in Hong Kong. After this virus began to circulate through the global human population, laboratories around the world began testing collected viruses to understand how they reacted to antibodies that were known to protect people from infections. As viruses immune to either antibodies or vaccines were discovered, clustered groups were eventually identified, and this led to early predictive modeling that helped vaccine manufacturers focus their efforts on emerging influenza lineages.
Co-author Justin Bahl, professor in UGA's College of Veterinary Medicine, said that these early research projects were instrumental in vaccine development.
"Clustering influenza viruses into specific groups to understand their level of immunity to vaccines was groundbreaking," he said. "However, this research was based on a virus surveillance methodology that is labor-intensive, time-consuming, and can be unreliable. We believed that a faster and more accurate method of predicting emerging seasonal influenza viruses could be accomplished using an infectious disease machine-learning algorithm."
The success of NIAViD opens up numerous avenues for further research, according to Regents' Professor John Drake, director of the Center for the Ecology of Infectious Diseases.
"Its ability to rapidly identify antigenic variants can support the ongoing development of flu vaccines, bolstering public health readiness and responsiveness to seasonal flu," he said. "Integrating NIAViD into surveillance systems would allow manufacturers to stay ahead of viral evolution, ensuring vaccines target the most current strains."
More information: Alpha Forna et al, Sequence-based detection of emerging antigenically novel influenza A viruses, Proceedings of the Royal Society B: Biological Sciences (2024). DOI: 10.1098/rspb.2024.0790