Clinical sequencing technology identifies new targets in diverse cancers
Novel abnormalities in the FGFR gene, called FGFR fusions, were identified in a spectrum of cancers, and preliminary results with cancer cells harboring FGFR fusions suggested that some patients with these cancers may benefit from treatment with FGFR inhibitor drugs, according to data published in Cancer Discovery, a journal of the American Association for Cancer Research.
FGFR genes are receptors that bind to members of the fibroblast growth factor family of proteins and play a role in key biological processes of a human cell. Because of a chromosomal abnormality, this gene sometimes fuses with another gene and forms a hybrid, or a gene fusion, resulting in a gene product with an entirely different function, causing cancers.
"We found targetable FGFR gene fusions across a diverse array of cancer types. Although rare for any individual cancer type, if found in an individual patient, these fusions are likely a major driver of that patient's cancer," said Arul M. Chinnaiyan, M.D., Ph.D., director of the Michigan Center for Translational Pathology at the University of Michigan in Ann Arbor. "We were surprised to find so many different FGFR fusions in so many different cancers.
"This study demonstrates the benefit of broad-based sequencing efforts in personalized oncology. It has the potential to identify novel, rare mutations that are 'actionable' therapeutic targets," Chinnaiyan added. "Such advances in sequencing technology facilitate rational precision therapies for individuals with late-stage cancer."
The Michigan Oncology Sequencing Program (MI-ONCOSEQ) facilitates integrative sequencing analysis of tumors from patients with advanced cancers. More than 100 patients have been enrolled since 2011. Through the project, researchers analyze the mutational landscape of each patient's tumor and suggest clinical trials or approved drugs that might be appropriate for that patient, according to Chinnaiyan.
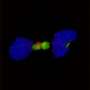
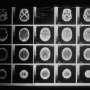
He and his colleagues identified novel fusions of the gene FGFR2 in the tumors of four patients recruited to MI-ONCOSEQ. Of these four patients, two had metastatic tumors of the bile duct; the third had metastatic breast cancer and the fourth had metastatic prostate cancer.
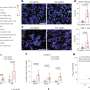
To further analyze whether FGFR fusions are present across different types of cancers, the researchers extended their assessment and analyzed data generated from an internal cohort of 322 patients, as well as from a large cohort of 2,053 patients recruited to The Cancer Genome Atlas. They identified several distinct FGFR fusions in nine different types of cancers, including bladder cancer, brain cancer and lung cancer.
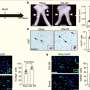
Chinnaiyan and colleagues then conducted studies using cancer cells and found that the different FGFR fusion proteins all seemed to drive cancer cell proliferation by activating FGFR signaling. The researchers were able to inhibit proliferation of the cells in vitro using the FGFR inhibitors PD173074 and pazopanib. In addition, they injected mice with human cancer cells and found that the tumors grown in mice could be inhibited with PD173074.
One of the four patients whose metastatic bile duct tumor failed to respond to conventional chemotherapy was recruited to an FGFR inhibitor trial, and he is currently undergoing treatment, according to Chinnaiyan.