Rice team rises to big-data breast cancer challenge
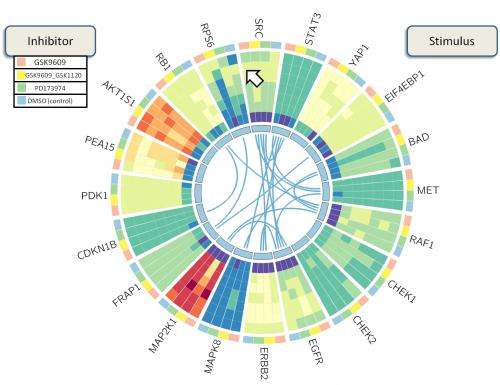
A colorful wheel developed by Rice University bioengineers to visualize protein interactions has won an international competition for novel strategies to study the roots of breast cancer.
The winning BioWheel by the Rice lab of bioengineer Amina Qutub was chosen this week, in the middle of Breast Cancer Awareness Month, topping 14 academic and industry participants in the HPN-DREAM Breast Cancer Network Inference Challenge. Qutub has been invited to present the lab's creation at the RECOMB/ISCB conference on Regulatory and Systems Genomics in Toronto next month.
The Rice team led by graduate student Wendy Hu won one of three subchallenges to create an intuitive, interactive tool to visualize "big data." In this case, that involved hundreds of thousands of data points about the effects of stimulators and inhibitors on protein networks drawn from a set of four breast cancer cell lines. Team members include postdoctoral researchers Byron Long and Dave Noren and undergraduate student Alex Bisberg.
All of the competitors were presented with the same data set. The idea, according to organizers, was to develop maps that increase the understanding of protein signaling in cancer cells and accelerate the development of treatments. Organizers of the crowdsource-style competition hoped that having dozens of participants work on the same data for four months would produce results that might otherwise take a single group many years.
"The task is to help people interpret intricate patterns that are very hard to see in high-dimensional data," said Qutub, an assistant professor of bioengineering based at Rice's BioScience Research Collaborative. Massive amounts of data can be produced when researchers study how healthy or cancerous cell lines taken from patients respond to stimulants, like drugs that up- or down-regulate protein interactions.
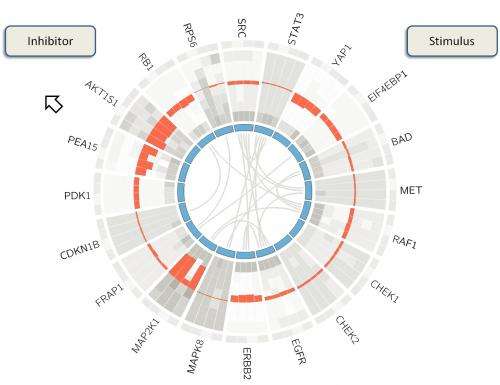
"BioWheel visualizes this data in a way that lets people quickly grasp changes in a protein and its connections over time," she said. "If you want to know how sets of proteins in a cancer cell change when you use a particular drug, this allows you to see their relationships quickly."
As presented in the summerlong competition, BioWheel showed connections between proteins and their expression levels as represented in colors that changed from light green (for no expression) to dark red (for high expression) as time progressed from the inside to the outside of the wheel. Shifting connections between proteins on the inner ring are seen in the center.
"The beauty of it is that it's a framework for all kinds of data sets," Qutub said, noting her lab will use BioWheel in an ongoing leukemia project. "We'll look not only at protein levels but also at genetic categories, patient age and gender to see how each parameter affects interactions in the cells."
The Qutub lab experiments on cell lines and builds computer models of protein-signaling networks that trigger such biological processes as cell growth, survival, death and migration. The researchers want to know how these networks operate in cancer cells to find more effective treatments for patients.
But BioWheel's design doesn't necessarily need to be applied to biological problems, Qutub said. "It could be just as effective to analyze interactions between political parties or links in an Internet network," she said. "It's for big data in general."
The Rice team also placed highly in a second subchallenge to reverse-engineer protein-interaction networks from the same set of cancer-cell data, along with a simulated dataset.
Qutub said the organizers are compiling a journal paper for submission and will incorporate BioWheel as part of a software package that will be available to researchers for free through the competition's nonprofit sponsors, Sage Bionetworks and the Dialogue for Reverse Engineering Assessments and Methods (DREAM), a collaboration between Columbia University and IBM.
In the meantime, the lab is working with Rice's Ken Kennedy Institute for Information Technology to continue the development of BioWheel and related network algorithms for compatibility with supercomputing resources, Qutub said.