Next-gen reappraisal of interactions within a cancer-associated protein complex
At a glance, DNA is a rather simple sequence of A, G, C, T bases, but once it is packaged by histone proteins into an amalgam called chromatin, a more complex picture emerges. Histones, which come in four subtypes—H2A, H2B, H3, and H4—can either coil DNA into inaccessible silent regions or untwist it to allow gene expression. To further complicate things, small chemical flags, such as methyl groups, affect whether histones silence or activate genes.
Among activator histones is a form of H3 decorated at a precise location (defined as H3K4) with three methyl groups (known as "H3K4me3"). Researchers knew previously that the presence of H2B exhibiting a single ubiquitin molecule stimulated the methylase that modifies H3K4, thereby increasing H3K4me3 levels. But how the methylase's activity was directed toward the appropriate targets had remained unclear.
Now, using biochemical, structural, and global sequencing techniques, researchers in the lab of Ali Shilatifard, Ph.D., an Investigator at the Stowers Institute for Medical Research, reveal an unanticipated mechanism underlying H3K4 trimethylation. Their study, published in the January 15, 2014 issue of Genes & Development, explains why H3K4me3 is deposited adjacent to a target gene promoter rather than haphazardly across the entire gene. This finding is significant because mutations in the human gene encoding the methylase responsible for H3K4me3 are associated with childhood leukemias, among other malignancies.
The work also illustrates the way powerful new genome-wide sequencing methodologies are impacting all molecular biology, including cancer research. "Here, we show that one cannot rely on methods that simply measure overall bulk H3K4me3 levels in vitro," says Shilatifard. "Only genome-wide sequencing could have revealed that H3K4 trimethylation was promoter-specific in non-mutant yeast."
This means that many assumptions about gene expression may need to be retested using next-generation approaches. "The old technologies were like observing just one region of the earth from a distant telescope in space and then making assumptions about what the entire earth looked like," Shilatifard says. "With new technologies, we can now see the whole planet."
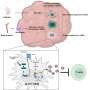
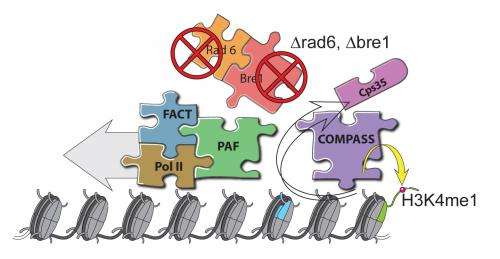
The methylase in question, named SET1 in yeast and MLL in mammals, is part of a protein aggregate called COMPASS, for COMplex of Proteins ASsociated with Set1. Shilatifard was the first to define the role of COMPASS in chromatin modification. "Over a decade ago, our lab used yeast to show that COMPASS was an H3 methylase," he says. "Since these fundamental systems are highly conserved from yeast to Drosophila to humans, we took advantage of the awesome power of yeast genetics to identify what regulates H3K4 methylation activity."
Part of the paper addresses SET1/MLL regulation by different proteins within yeast COMPASS. Investigators knew that if you lopped off more than half of SET1's front end, levels of DNA-bound trimethylated H3K4 in cells harboring the remaining "stub" were equal to those in cells containing the full-length protein when analyzed in bulk. This led some to presume that the entire front end of SET1/MLL, as well as factors that interact with it, must not be needed to regulate H3K4me3 activity.
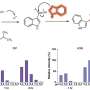
The new paper shows that this presumption was not correct. The Shilatifard team first employed biochemical methods to capture every piece of DNA bound to H3K4me3 in the genome of yeast harboring either full-length SET1 or the stub missing the front end. They then sequenced all of those DNA fragments and mapped their position in the yeast genome.
Significantly, they found that even though H3K4me3 levels in bulk were equivalent in normal and mutant cells, H3K4me3 was differentially distributed throughout the genome: in normal cells, H3K4me3 complexes sat primarily on DNA regions that switch adjacent genes on or off, control regions called promoters. By contrast, the DNA of cells harboring the stub exhibited DNA-binding H3K4me3 complexes in the middle of or between genes.
These discoveries, combined with other findings, call for re-interpretation of data suggesting that the stub is all you need for H3K4 trimethylation. Instead, the new work shows that COMPASS factors that bind to the SET1/MLL front end limit H3K4me3 deposition to the correct genomic sites (that is, to the promoter regions), while factors that bind the SET1/MLL stub increase the protein's half-life. This partially explains earlier misinterpretations: highly stable stubs of SET1 "promiscuously" methylated the wrong parts of the genome when the regulatory front end of the protein was missing. The paper also addressed how H2B ubiquitin modification machineries stimulate the entire process.
Understanding COMPASS regulation is essential, as genes encoding factors in the complex are mutant in numerous cancers. For example, chromosomal translocations involving a gene encoding one MLL protein occur frequently in human leukemias, hence the designation MLL, which stands for Mixed Lineage Leukemia protein. Other MLL proteins are strongly implicated as tumor suppressors in human cancers such as lymphoma and pediatric brain tumors.