Pinpointing long range genomic connections to determine the genetic basis of disease
Researchers at the Babraham Institute and the Francis Crick Institute have developed and used a new technique to join the dots in the genomic puzzle. Just as a dot to dot puzzle needs to be completed to visualise the full picture, the researchers' analysis connected regulatory elements called promoters and enhancers and showed their physical interactions over long distances within the mouse and human genomes. The ability to map promoter-enhancer interactions in the human genome has huge potential in understanding the genetic basis of disease.
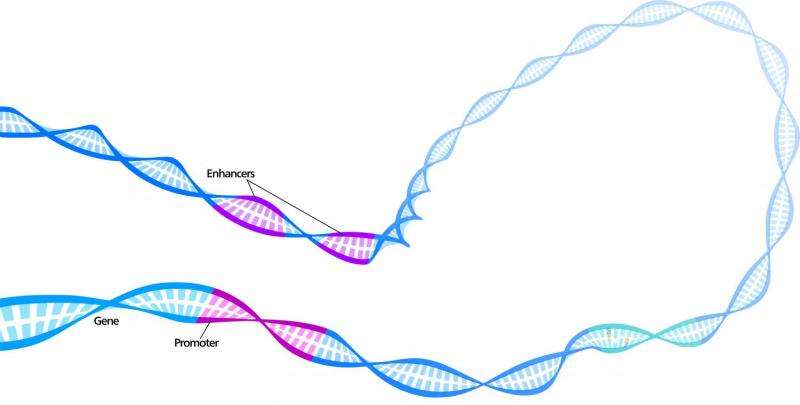
Our development as an embryo and the establishment of different cell types in the body is dependent on a suite of genomic regulatory elements to orchestrate the correct expression of genes in different locations and at different times. To have a complete understanding of how a gene is regulated, both in health and disease, it is necessary to have a comprehensive catalogue of the regulatory elements that contribute to its control.
The Babraham Institute researchers refined an existing technique to look at the one million regulatory elements in the mouse genome and link these to gene promoters to understand how genes are switched on and off. At the same time, the technique was used to study human blood cell types. If the genome is imagined as a linear stretch of DNA sequence, the research pinpointed sections of the genome where it loops to bring regulatory elements controlling gene expression into physical contact with each other. In genomic distances, enhancer regions can be hundreds of kilobases of DNA letters (1KB is 1000 letters or bases) away from the genes they regulate.
Previous interaction assays weren't able to provide sufficient resolution to link regulatory elements with specific promoters. To solve this problem, the team at the Babraham Institute came up with a clever solution: using RNA 'baits' to be able to pull out just the genomic fragments containing promoters from the melting pot of a hundred billion (1011) genomic interactions in the mouse genome. The technique is called Promoter Capture Hi-C. This research, published online in the journal Genome Research in March, served as a proof of principle for the use of Promoter Capture Hi-C to map genomic interactions in mouse cells at high resolution. The human cell analysis now published in Nature Genetics by researchers from the Francis Crick Institute, the Babraham Institute and King's College London presents the most extensive genome-wide map of promoter-enhancer interactions in the human genome.
Dr Peter Fraser, Head of the Nuclear Dynamics programme of research at the Babraham Institute and senior author on the Genome Research paper reporting this work said: "These results provide the first genome-wide catalogue of interactions between gene promoters and their long-range interacting elements. Previous methods were akin to analysing a bucket of seawater and using this to make assumptions on the ocean's contents. With Promoter Capture Hi-C we can trawl for specific physical associations between regulatory elements that control gene expression, and use this information to build up a more complete picture of the genome's three-dimensional shape to help us understand how this functions in health and disease."
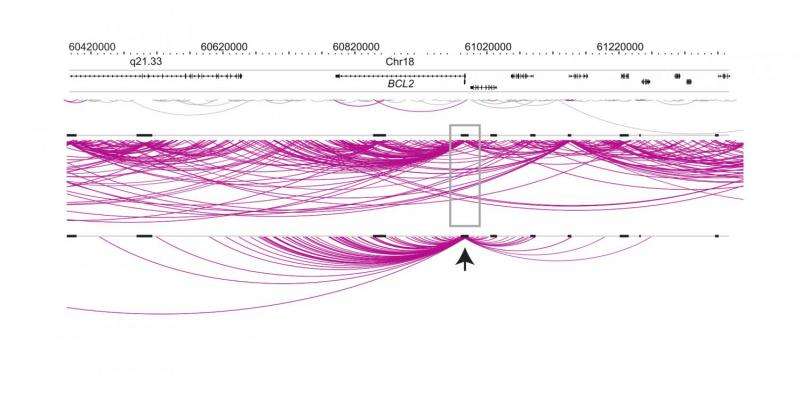
Use of the Promoter Capture Hi-C technique to delve the human genome pinpointed the long-range interactions of nearly 22,000 promoters, identifying millions of interactions and providing an unprecedented snapshot of the distal genomic regions that contact promoters. Genome-wide association studies have uncovered thousands of specific areas of the genome (loci) that have been shown to be associated with different diseases, including within regulatory regions. Knowing which genes a regulatory region affects has so far been extremely difficult and this has been a major roadblock to understanding genome-wide association studies. The resolution allowed by Promoter Capture Hi-C showed that the regions that interact with promoters are highly enriched for DNA mutations (SNPs; single nucleotide polymorphisms) that have been associated with disease and means that researchers can now link potentially defective regulatory elements of the genome with the genes they influence.
Dr Cameron Osborne, from King's College London but who undertook this research while at the Babraham Institute, senior author on the Nature Genetics paper, said: "Our data physically ties the GWAS SNPs to putative gene targets, and shows that they commonly interact with more distal genes rather than the nearest neighbours. The identification of GWAS target genes has the potential to unleash a new phase of characterising polymorphisms and the genes and molecular pathways they affect."
In addition to linking regulatory elements to active genes, the analysis in human cells also identified connections with inactive genes and elements that appear to function as transcriptional silencers. A lot less is known about what switches genes off compared to our understanding of what switches them on. The characterisation of such elements may help to define a genomic signature for silencer elements, allowing them to be more easily identified throughout the genome. Ultimately, this may shed light on the mechanisms which suppress gene expression.
More information: Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C, DOI: 10.1038/ng.3286
Schoenfelder, Furlan-Magaril, Mifsud et al. (2015). The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Research, genome.cshlp.org/content/25/4/582.full