Tapping the genome's social network to find cancer drivers
Any one tumor might harbor mutations in thousands of different genes. The challenge is to find the driver mutations—which fuel cancerous activity, and might be promising treatment targets—within the haystack of passengers (mutations that, while present in the tumor, do not help it grow or spread).
In a paper in Nature Methods, a team led by Heiko Horn and Kasper Lage of the Broad Institute's Stanley Center for Psychiatric Research and Massachusetts General Hospital MGH; Michael Lawrence and Jesse Boehm of the Broad Cancer Program; and Gad Getz of the Cancer Program and MGH describe NetSig, an open-source computational tool that looks for cancer-driving mutations by joining cancer genome data with protein interaction data. NetSig is designed to complement existing tools and expand discovery from cancer genomes in any existing analysis pipeline.
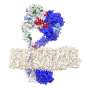
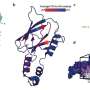
Proteins interactions reveal a sort of genomic Facebook, a social network through which genes share information and carry out a cell's functions. The added functional perspective can help researchers identify drivers with greater confidence, especially in genes that mutate only rarely.
To that end, NetSig taps InWeb_InBioMap (InWeb_IM), a protein interaction map with data on more than half a million protein-protein interactions. Lage's lab developed InWeb_IM in 2016 for functionally interpreting large genomic data sets.
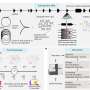
To develop NetSig, the team merged InWeb_IM with The Cancer Genome Atlas (TCGA)-derived exome data from 4,742 tumors spanning 21 cancer types. The team then tested the tool's capabilities in a series of in silico and in vivo experiments, finding that NetSig could:
- Recognize known cancer drivers in 60 percent of tested tumor types, including those with relatively few samples.
- Predict new driver genes that validation experiments—conducted with the Cancer Program's Target Accelerator initiative—showed were truly tumor-promoting.
- Identify hitherto-unnoticed drivers lurking in TCGA and other datasets. In particular, NetSig revealed that between 4 and 14 percent of lung tumor patients previously deemed oncogene-negative (that is, had no detectable oncogenes present) may actually harbor cancer-promoting extra copies of the genes AKT2 or TFDP2.
More information: Heiko Horn et al. NetSig: network-based discovery from cancer genomes, Nature Methods (2017). DOI: 10.1038/nmeth.4514
Taibo Li et al. A scored human protein–protein interaction network to catalyze genomic interpretation, Nature Methods (2016). DOI: 10.1038/nmeth.4083