This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Taking a 3D view of the genome may help treat pediatric brain cancers
Researchers led by Sanford Burnham Prebys assistant professor Lukas Chavez, Ph.D., are leveraging the latest technology to take a never-before-seen look at ependymoma, one of the deadliest pediatric brain tumors. By visualizing how the genome is organized and arranged within tumor cells, they were able to reveal genes in tumors that may be future targets for therapy. The results appear in Nature Communications.
"The human genome is made up of many protein-coding genes and an even greater number of noncoding sections, which are all tightly packaged and coiled up to fit inside the tiny nucleus of a cell," says Chavez. "We're using cutting-edge technologies to look at the way that genome is packaged and coiled, which gives us a unique perspective on the mechanisms of gene regulation. This approach helps us understand the link between the shape of the genome and cancer."
Tumors of the brain and spinal cord, including ependymomas, are the most common malignant cancers in children up to age 14 and the leading cause of death by disease during childhood in the United States.
"Ependymomas come in many different genetic and molecular subtypes that can affect how well patients respond to treatment," says Chavez. "The current standard-of-care treatment includes surgery followed by radiation, which bears the risk of long-term, therapy-induced neurological side effects as well as secondary cancers. New, targeted therapeutic options for ependymoma are desperately needed. If successful, our research will lead to new, effective medications to treat these dangerous cancers."
The researchers focused on the most common and aggressive forms of the disease, which occur mainly in young children. They used an emerging technique called 3D genome mapping, which visualizes how genes are organized within the nucleus of cells.
"Science has historically studied the genome in two dimensions, focusing on how genes are arranged in long, linear sequences," says Chavez. "But the genome is a 3D object like anything else, and the way that genes are arranged in space makes a difference in how those genes are expressed in the body."
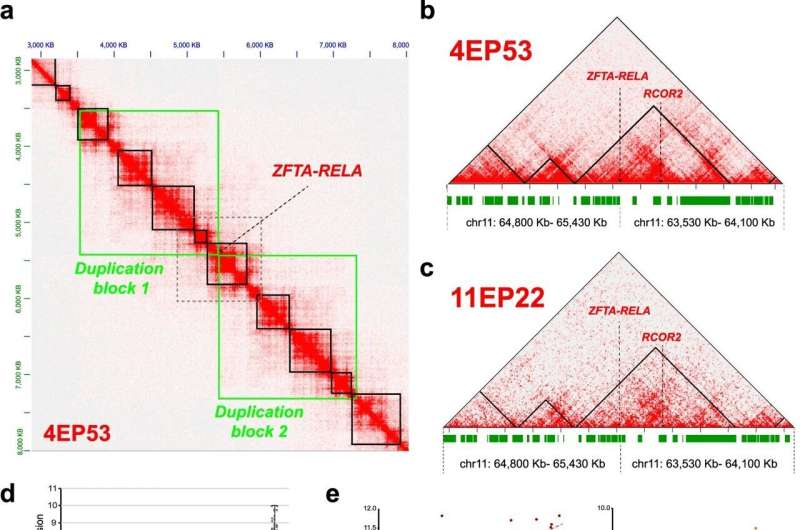
The researchers studied previously undiscovered loops in the genomes of ependymoma tumors. These loops change the way that genes are expressed, which in turn produces signals that help tumors grow.
"We've confirmed that configuration of these genes in the loops are essential for ependymoma tumors," says Chavez. "This means that we now have a host of new candidate targets for treatments that we would never have been able to identify without this technology."
The researchers believe that their results will lay the foundation for further studies that will lead to future therapies. The researchers are also planning to look at other pediatric cancers, since many of these diseases have few therapeutic options.
"There are alarming numbers of tumors that we struggle to treat because we simply don't know how they work from a biological standpoint," says Chavez. "This work shows that there's a lot more we still don't know about the genomics of tumors, and unlocking these mysteries may be the key to finally overcoming these aggressive cancers."
More information: Konstantin Okonechnikov et al, 3D genome mapping identifies subgroup-specific chromosome conformations and tumor-dependency genes in ependymoma, Nature Communications (2023). DOI: 10.1038/s41467-023-38044-0