This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
A human protein found in non-immune cells defends against COVID-19
Earlier this month, health authorities confirmed that BA.2.86, a highly mutated variant of the virus that causes COVID-19, is at risk of spreading rapidly in countries around the world. With updated shots rolling out this week across the United States, it remains to be seen whether this newest vaccine campaign will prove effective in slowing the spread of BA.2.86 or other emerging variants.
Still, vaccines remain the most powerful therapeutic tool to prevent and slow the spread of COVID-19. But, recent research by Howard Hughes Medical Institute Investigator John MacMicking and colleagues at Yale University now published in Nature found that a defense protein in cells that line the lungs and other non-immune tissues could one day offer additional treatment pathways, particularly for those vulnerable to severe COVID-19 infection.
In humans, the immune system—a complex network of organs, proteins, and cells—works collectively to mount a protective response against outside invaders such as viruses and bacteria. When scientists refer to the cells that make up the human immune system, they traditionally mean cells that originate in the bone marrow, such as B cells, T cells, macrophages, and dendritic cells.
But, in reality, the immune system does not operate as a closed system in the human body, rather, it permeates every part of our physiology. Cells that have little or no connection with the bone marrow—such as the epithelium lining the airways or digestive tracts, hepatocytes comprising the liver, or neurons transmitting signals within the brain—can also play a crucial role in combatting viruses or other pathogens as part of the cell-autonomous innate immune system.
Unlike antibodies, these mechanisms do not often "remember" specific pathogens they have encountered but, they can respond to instructions from T-cells, which do have immunological memory. Knowing that the first encounter with SARS-CoV-2 typically occurs within the respiratory tract, MacMicking and his collaborators searched for ways to trigger lung epithelial cells to wage war against the virus, independent of the body's antibody response.
"Protective immunity to SARS-CoV-2 not only relies on antibodies generated by vaccination or during the infection itself, but also by turning on host defense proteins inside infected cells to help bring infection under control," says MacMicking. "That occurs all over the body: in the airways, of course, but also in many target tissues. Indeed, cells that are not normally considered a part of the immune system are often instructed by it to produce local defense proteins that can directly inhibit SARS-CoV-2."
Vaccines to protect against COVID-19 or other viruses work by generating neutralizing antibodies capable of binding to the virus when a person becomes infected. Over time, as viruses spread and mutate, vaccines can struggle to keep pace with the emergence of new strains that evade antibody neutralization. Moreover, in the case of COVID-19, it is still unclear why some individuals experience mild disease symptoms—or no symptoms at all—whereas others become seriously ill.
A new pathway to halt replication
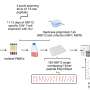
Recognizing these challenges, MacMicking and his collaborators probed the human genome in search of new pathways that interfere with how the SARS-CoV-2 virus replicates inside human cells, most specifically, within cells in our airways.
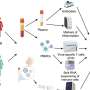
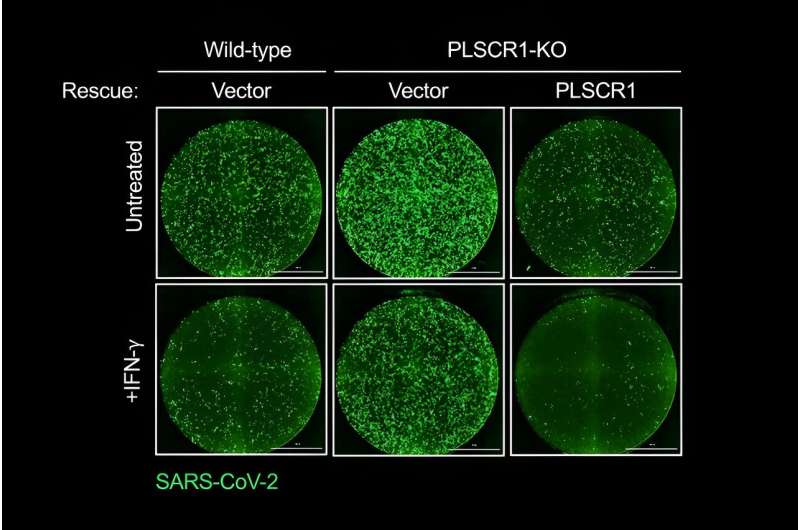
The research team discovered that cells that line our lungs and other tissues express a protein, phospholipid scramblase 1 (PLSCR1), that can halt SARS-CoV-2 replication in its tracks before the virus spreads to nearby cells. This protein is expressed in cells before infection but it kicks into gear when activated by interferons (IFNs), a group of cytokines that instructs cells to defend against foreign invaders. This type of local cell-autonomous immunity plays an important role in safeguarding mucosal barriers and target tissues against deadly pathogens, including those that cause tuberculosis, typhoid, and AIDS.
With this knowledge, MacMicking and his team set out to investigate whether they could tap cell-autonomous immunity to combat SARS-CoV-2.
"Scientists don't really know a lot about the non-classical immune system—that is, cell populations, which originate outside of the bone marrow, and the roles they play in protecting against infection," says MacMicking. "Our lab is working to advance understanding of these understudied cell types, which, historically, have not been considered part of the immune system. Indeed, just as immune cells participate in the homeostasis of every organ system, the opposite may also be true—cells of every organ contribute to immunity, including direct defense against infection."
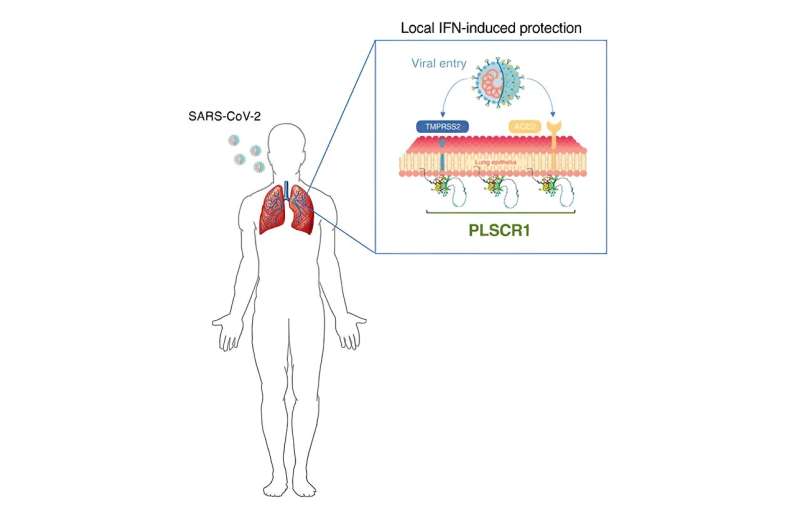
In addition to helping redefine the boundaries of our immune system, MacMicking and his team's study of PLSCR1 might one day point to new therapeutic strategies to treat or prevent COVID-19. Such treatments could be deployed in conjunction with vaccines or other antibody-based therapies, especially against newer SARS-CoV-2 strains that exhibit evasion to some antibody types.
"This is a beautiful piece of work that teaches us more about the intricate defenses hard-wired into our cells that protect us from infection and disease," says Charles Rice, a professor of virology at Rockefeller University. "A deeper mechanistic understanding of how PLSCR1 blocks SARS-CoV-2 infection may reveal new angles for developing broadly effective antivirals."
MacMicking and his team's findings also offer hope for severe COVID-19 patients who may be unresponsive to treatment with convalescent plasma or remdesivir. Here, pilot studies have shown that IFN therapy can provide another treatment avenue. Previous studies from the lab of HHMI Investigator Jean-Laurent Casanova at Rockefeller University have shown a strong connection between severe COVID-19 and individuals who have genetic mutations that inhibit interferon signaling.
"If we can find ways to turn on the PLCSR1 gene in individuals with an interferon signaling deficiency, or bypass the need for such signaling altogether, then it could be a useful way to think about new therapeutic interventions against SARS-CoV-2," says MacMicking. In addition, by screening patients for PLSCR1 mutations, doctors could potentially identify individuals who face a high risk of experiencing severe COVID-19 symptoms.
While the findings by MacMicking and his collaborators are compelling, they are just a first step in the search for new therapeutic options to treat COVID-19 and other viruses.
One of the biggest obstacles ahead for the research team is that IFNs can activate hundreds of different host defense proteins, including those which may not be beneficial for the clinical course of COVID-19. "Knowing which proteins are useful and which proteins are not allows us to take steps toward developing a small molecule or drug that can potentially mimic [PLSCR1's] actions without turning on IFN-induced proteins that may be detrimental for the host," MacMicking says.
The group's next task is to map the native structure of the protein, he adds. "Once we know exactly what the protein looks like at the atomic level, we can make better predictions about how it works to block SARS-CoV-2, which may lead to the design of drugs that promote its activity."
Although this work principally focuses on SARS-CoV-2, the group's research could have broader implications for how therapeutic preventions and treatments are designed for other viruses.
"When it comes to human health, we don't have that many antiviral drugs, unlike the case for bacteria, where our antibiotic war chest is historically much larger," says MacMicking. "Today, our antiviral response options consist primarily of vaccines and nucleoside or polymerase inhibitors that have some limitations.
"With this in mind, one aim [of our group's work] is to expand our ability to combat viruses. If we can start to think about ways to appropriately turn on local or tissue-resident immunity—and especially the antiviral proteins involved—then we can try to design chemicals or drugs that allow us to better activate those pathways."
More information: Dijin Xu et al, PLSCR1 is a cell-autonomous defence factor against SARS-CoV-2 infection, Nature (2023). DOI: 10.1038/s41586-023-06322-y