This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
Proteomic quantification of native and ECM-enriched mouse ovaries reveals an age-dependent fibro-inflammatory signature
A new priority research paper titled "Proteomic quantification of native and ECM-enriched mouse ovaries reveals an age-dependent fibro-inflammatory signature" has been published in Aging.
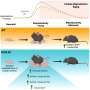
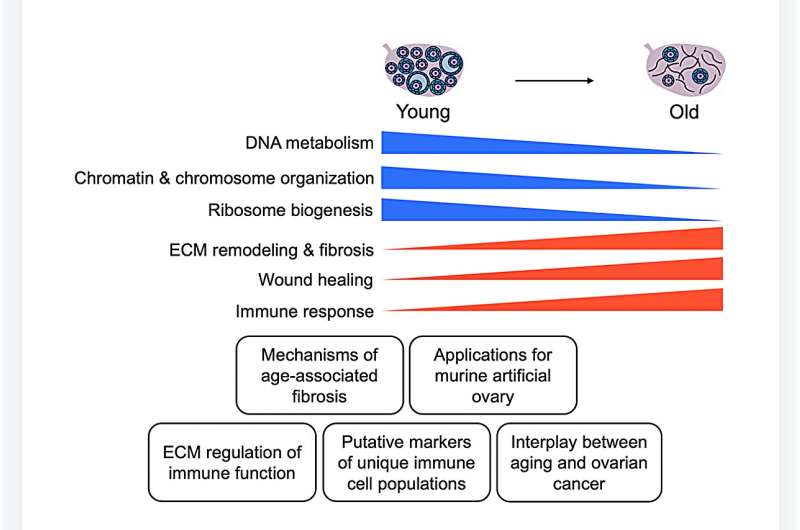
The ovarian microenvironment becomes fibrotic and stiff with age, in part due to increased collagen and decreased hyaluronan. However, the extracellular matrix (ECM) is a complex network of hundreds of proteins, glycoproteins, and glycans which are highly tissue-specific and undergo pronounced changes with age.
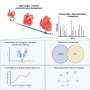
In this new study, researchers Shweta S. Dipali, Christina D. King, Jacob P. Rose, Joanna E. Burdette, Judith Campisi, Birgit Schilling, and Francesca E. Duncan from Northwestern University's Feinberg School of Medicine, the Buck Institute for Research on Aging and the University of Illinois at Chicago used label-free quantitative proteomic methods to define comprehensive, age-dependent changes in the murine ovarian proteome and ECM in an unbiased manner.
"To obtain an unbiased and comprehensive profile of age-associated alterations to the murine ovarian proteome and ECM, we used a label-free quantitative proteomic methodology," the researchers explain.
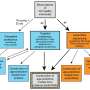
The researchers validated conditions to enrich for the ECM prior to proteomic analysis. Following analysis by data-independent acquisition (DIA) and quantitative data processing, they observed that both native and ECM-enriched ovaries clustered separately based on age, indicating distinct age-dependent proteomic signatures. The team identified a total of 4,721 proteins from both native and ECM-enriched ovaries, of which 383 proteins were significantly altered with advanced age, including 58 ECM proteins.
Several ECM proteins upregulated with age have been associated with fibrosis in other organs, but to date their roles in ovarian fibrosis are unknown. Pathways regulating DNA metabolism and translation were downregulated with age, whereas pathways involved in ECM remodeling and immune response were upregulated. Interestingly, immune-related pathways were upregulated with age even in ECM-enriched ovaries, suggesting a novel interplay between the ECM and the immune system.
Moreover, the researchers identified putative markers of unique immune cell populations present in the ovary with age. These findings provide evidence from a proteomic perspective that the aging ovary provides a fibroinflammatory milieu, and their study suggests target proteins which may drive these age-associated phenotypes for future investigation.
"To our knowledge, this is the first study to utilize unbiased proteomic approaches to investigate the effect of reproductive aging on the murine ovarian proteome and matrisome," the researchers conclude.
More information: Shweta S. Dipali et al, Proteomic quantification of native and ECM-enriched mouse ovaries reveals an age-dependent fibro-inflammatory signature, Aging (2023). DOI: 10.18632/aging.205190