This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
New approaches needed to find better cancer drug targets, says cancer specialist
Tumors depend heavily on certain genetic changes to thrive, and researchers have discovered many such "genetic dependencies" as targets for potential new cancer drugs. At the Broad Institute of MIT and Harvard and elsewhere, researchers are also learning how dependencies affect cancer cells and how they influence each other and contribute to drug resistance.
But the field needs fresh approaches to finding dependencies that could lead to new classes of more effective, even curative, cancer drugs, says Bill Sellers, who is director of the Broad's Cancer Program. "We don't often think hard enough about how to get to cures in cancer," said Sellers, who is also a core institute member at Broad and faculty member and senior advisor to the president for experimental therapeutics at Dana-Farber Cancer Institute.
Since 2018, the Cancer Dependency Map (DepMap) Consortium, an academic-industrial partnership launched by the Broad, has uncovered several potential drug targets by systematically screening cancer models in search of genetic dependencies.
As part of the Cancer Program at the Broad, the Sellers lab explores the link between genetic alterations and cancer dependencies with the goal of informing new therapies.
Sellers sat down with us to talk about the limitations of current approaches in cancer dependency research and what is needed to move closer to cures.
How have researchers looked for cancer dependencies in the past?
For the most part, people have knocked out one gene at a time at the genome scale and asked, "What's the consequence of losing that gene's function for the growth of cancer?"
Initially, there were two approaches: small hairpin RNA (shRNA)-based and now CRISPR-based experiments. There are a number of limitations to those approaches.
First of all, they're completely in vitro, so they don't take into consideration any of the factors that might be provided in the tumor microenvironment or the host. That's been difficult to address.
Cancer models are also a limitation. You can only do these studies if you have cell lines or models that can be manipulated in vitro at the genome scale. There are a whole host of cancers and cancer subtypes for which we don't have many cell lines. For example, in prostate cancer, there are maybe three common cancer cell lines, even though it's one of the most common diseases. Usually people attempt to establish models by in vitro selection or by taking biopsies from patients, which is long and laborious.
What new kinds of dependencies are you interested in?
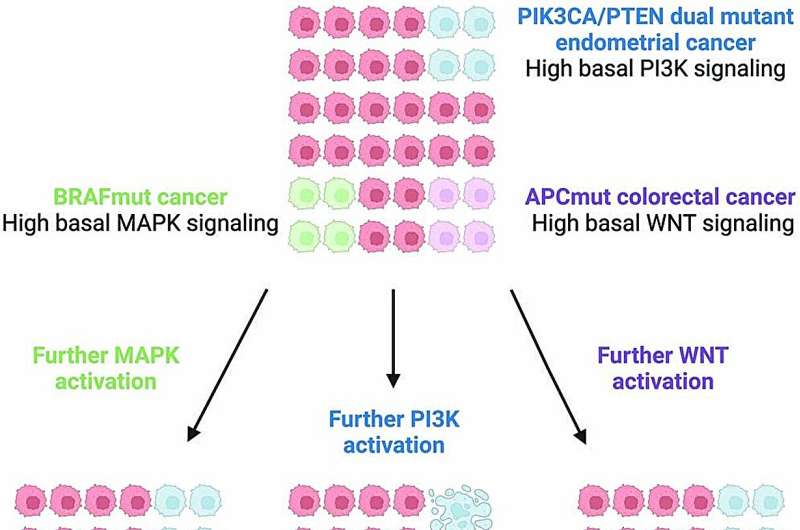
Over time, evidence has emerged for the idea of a Goldilocks zone for cancer—that tumors may need an optimal amount of signaling or "gas" to fuel their growth. To maintain this optimal signaling, cancers likely modulate gene activity by not just increasing but also decreasing gene expression. In a paper we published in September in Nature Genetics, we focused on the idea that cancers might be susceptible not just to loss of function of certain genes or gene products, i.e. having too little gas, but also gain of function, or too much gas.
We've also been focusing on the limitation of studying one gene at a time when it's possible that two genes are performing a similar enough function. You'd need to disable both genes to see how important they are as a drug target. This is the case for highly related paralogs, which are genes with a common genetic ancestor and similar functions. A drug might need to target both paralogs to produce a therapeutic effect. We miss that combinatorial dependence in our current screens.
A few years ago, we were building a CRISPR library targeting pairs of closely related genes and we thought we'd study NRAS-mutant melanoma. But we discovered two negative regulators [inhibitors of gene expression], DUSP4 and DUSP6. When they were both depleted, we saw ERK hyperactivation and loss of viability of the NRAS-mutant tumors. We found that cancers could be adversely affected both by inhibitors and activators of that pathway.
Perhaps most exciting, Cory Johannessen (former Broad scientist, now at Novartis) took drug-resistant cells and looked for single-gene knockouts that were increasing in their dependency compared to the non-resistant cells. As the cells were becoming resistant, their dependency on negative regulators became higher and higher.
This is quite exciting because most of the time, we're trying to create two drugs that have completely separate resistance mechanisms. But in this case, it looks like a first drug or inhibitor would elicit a mechanism of resistance that would increase the cancer's dependency on the second druggable node.
This would mean that for these two mechanisms, drug combinations could be given in cycles, sequentially, rather than together at the same time in order to exploit the nature of the evolution of the cancer. It suggests that we're missing this entire class of potential therapeutics. This is something we're testing right now.
Where do you think the search for genetic dependencies in cancer needs to go in the future, and what tools will that entail?
In my first go-around I thought, "We're going to solve all of cancer in one fell swoop." I think the reality is: Each new experimental approach that we take will solve some piece of the puzzle. Cancer is extraordinarily complex and difficult, and we have to tackle these in doable blocks.
If we want to exploit gain-of-function, then we need to have gain-of-function perturbations. Right now we have CRISPR activation (CRISPRa), which turns on genes but not a mutated, activated form of the gene. That could be achieved potentially by something like CRISPR editing.
The combinatorial problem is also extremely significant, especially if the reagents you use are not all validated. Having highly validated knockout reagents for every gene would help narrow down the number of reagents that you need to do effective combinatorial screens.
What progress do you hope to see in this field in the next five years?
We don't often think hard enough about how to get to cures in cancer. We know that it's going to require combination therapeutics, whether that's combinations of inhibitors or sequential treatment with inhibitors and activators. In thinking about how to get there, a lot of our attempts don't really address the fundamental issue of resistance.
I think this idea of taking drugs that we know work, deeply understanding how cancers develop resistance to those drugs, and rationally understanding the common functional consequences of these resistance mutations, is one of our best chances to get to combination therapies that have a chance at curing patients.
More information: Liang Chang et al, Systematic profiling of conditional pathway activation identifies context-dependent synthetic lethalities, Nature Genetics (2023). DOI: 10.1038/s41588-023-01515-7