This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Study of different autism types finds shared mechanism that may respond to drugs

An analysis of how brains with different forms of autism develop has revealed common underlying mechanisms that may respond to existing medications.
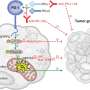
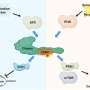
For the study, Rutgers Health researchers used a technique called induced pluripotent stem cells to transform the blood cells of people with both genetic and unexplained (or idiopathic) autism spectrum disorder (ASD) into early brain cells called neural precursor cells.
As the precursor cells from both groups matured in the lab, defects in a common signaling pathway that controls structural proteins led them to struggle with an important step in cell differentiation, the growth of neurites, and the cell migration needed for proper brain architecture.
Although some cell lines exhibited too much activity in this mTOR pathway, while others exhibited too little, the researchers could correct both problems and spur better cell differentiation with existing drugs that either stimulate or inhibit the activity of mTOR (mechanistic target of rapamycin).
"Cells in a dish are not fully human cells that have developed in a fetus and functioned in a person, but they are a lot closer than mouse cells," said Emanuel DiCicco-Bloom, a professor of neuroscience and cell biology/pediatrics at Robert Wood Johnson Medical School and senior author of the study in eLife.
"This finding is particularly interesting because the process of growing new synaptic spines when people learn things is completely analogous to the processes we observed in the cells we used for this experiment: growing axons and migrating during fetal development," DiCicco-Bloom said.
"So even though this experiment mimicked a process you'd see during early to mid-pregnancy, the same process involving structural proteins is happening right now in you and me, which means that if we took cells from people with autism and found this abnormal regulation of mTOR in their cells in a dish, those people might be candidates as adults for mTOR regulating drugs to improve their function."
The visible symptoms of ASD vary widely but typically feature some repetitive behaviors and some impairment in communication and social interaction. The condition's incidence has increased from about 1 in 150 children in 2000 to 1 in 36 children in 2020, according to the Centers for Disease Control and Prevention. Roughly 10 to 15 percent of people with ASD have genes that are known to elevate their risk for ASD. Other cases are idiopathic, meaning they are unexplained.
Rutgers Health researchers began the study with blood from three unrelated people with idiopathic ASD, ages 4 to 14 years, with the expectation of finding person-specific differences in the processes occurring during development in utero.
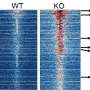
When the researchers used the pluripotent stem cell technique to transform blood cells into the sort of neuron precursors typically found in fetal brains, they unexpectedly found many similarities, including abnormalities in the mTOR pathway, which regulates cell creation, metabolism, neurite growth, remodeling, and destruction, among many other functions.
The researchers then gained access to blood cells from another three patients with ASD caused by a particular genetic abnormality associated with about 1 percent of ASD, deletion of genes on chromosome 16, called 16p11.2 deletion. They performed the same experiment and found the same disruptions in neuron development.
Subsequent analysis showed that the mTOR signaling disruptions in some patients stemmed from excessive amounts of a particular molecule, while the disruptions in others stemmed from insufficient amounts. In either case, the researchers could use existing medications approved for use in other conditions to correct the problem and stimulate normal development.
The study team has already begun a follow-up investigation to see if people with ASD stemming from other genetic causes exhibit similar disruptions in mTOR activity during development. If mTOR signaling disruption proves a common feature of ASD, tests of mTOR function could help clinicians diagnose the condition more accurately and differentiate it from other conditions with similar effects.
"A couple of very rare genetic types of autism have already been linked to the mTOR pathway, but this is the first to connect mTOR with genes in the 16p11.2 area, which does not have mTOR on it, and with three presumably different types of idiopathic autism from three unrelated people," said Smrithi Prem, lead author of the study and a psychiatry resident at Penn Medicine who led the study as an MD/Ph.D. student at Robert Wood Johnson Medical School.
"These findings also echo something that has appeared in studies of other conditions, that not all people with mTOR dysregulation have excessive activation that needs inhibition," Prem said.
"There are two kinds of mTOR dysregulation, but most trials we've run on people with mTOR dysregulation have only used inhibitors. Our findings showed that cells from two of the people we studied needed more mTOR, not less, and that may spur trials that give different types of mTOR treatment to different individual patients."
More information: Smrithi Prem et al, Dysregulation of mTOR signaling mediates common neurite and migration defects in both idiopathic and 16p11.2 deletion autism neural precursor cells, eLife (2024). DOI: 10.7554/eLife.82809