This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Scientists leverage machine learning to decode gene regulation in the developing human brain
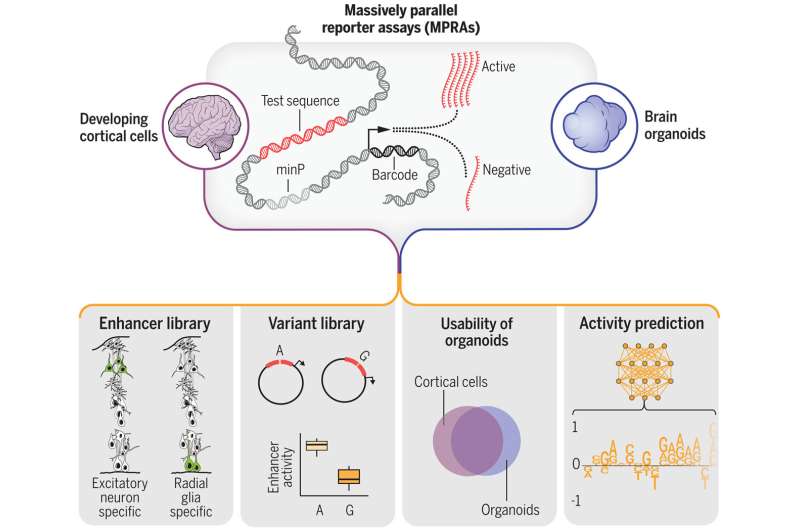
In a scientific feat that broadens our knowledge of genetic changes that shape brain development or lead to psychiatric disorders, a team of researchers combined high-throughput experiments and machine learning to analyze more than 100,000 sequences in human brain cells—and identify over 150 variants that likely cause disease.
The study, from scientists at Gladstone Institutes and University of California, San Francisco (UCSF), establishes a comprehensive catalog of genetic sequences involved in brain development and opens the door to new diagnostics or treatments for neurological conditions such as schizophrenia and autism spectrum disorder. The paper, "Massively Parallel Characterization of Regulatory Elements in the Developing Human Cortex," appears in the journal Science
"We collected a massive amount of data from sequences in noncoding regions of DNA that were already suspected to play a big role in brain development or disease," says Senior Investigator Katie Pollard, Ph.D., who also serves as director of the Gladstone Institute for Data Science and Biotechnology.
"We were able to functionally test more than 100,000 of them to find out whether they affect gene activity, and then pinpoint sequence changes that could alter their activity in disease."
Pollard co-led the sweeping study with Nadav Ahituv, Ph.D., professor in the Department of Bioengineering and Therapeutic Sciences at UCSF and director of the UCSF Institute for Human Genetics. Much of the experimental work on brain tissue was led by Tomasz Nowakowski, Ph.D., associate professor of neurological surgery in the UCSF Department of Medicine.
In all, the team found 164 variants associated with psychiatric disorders and 46,802 sequences with enhancer activity in developing neurons, meaning they control the function of a given gene.
These "enhancers" could be leveraged to treat psychiatric diseases in which one copy of a gene is not fully functional, Ahituv says, "Hundreds of diseases result from one gene not working properly, and it may be possible to take advantage of these enhancers to make them do more."
Organoids and machine learning take the spotlight
Beyond identifying enhancers and disease-linked sequences, the study holds significance in two other key areas.
First, the scientists repeated parts of their experiment using a brain organoid developed from human stem cells and found that the organoid was an effective stand-in for the real thing. Notably, most of the genetic variants detected in the human brain tissue replicated in the cerebral organoid.
"Our organoid compared very well against the human brain," Ahituv says. "As we expand our work to test more sequences for other neurodevelopmental diseases, we now know that the organoid is a good model for understanding gene regulatory activity."
Second, by feeding massive amounts of DNA sequence data and gene regulatory activity to a machine learning model, the team was able to train the computer to successfully predict the activity of a given sequence. This type of program can enable "in-silico" experiments that allow researchers to predict the outcomes of experiments before doing them in the lab. This strategy enables scientists to make discoveries faster while using fewer resources, especially when large quantities of biological data are involved.
Sean Whalen, Ph.D., a senior research scientist in the Pollard Lab at Gladstone and a co-first author of the study, says the team tested the machine learning model using sequences held out from model training to see if it could predict the results already gathered on gene expression activity.
"The model had never seen this data before and was able to make predictions with great accuracy, showing it had learned the general principles for how genes are impacted by noncoding regions of DNA in developing brain cells," Whalen says. "You can imagine how this could open up a lot of new possibilities in research, even predicting how combinations of variants might function together."
A new chapter for brain discoveries
The study was completed as part of the PsychENCODE Consortium, which brings together multidisciplinary teams to generate large-scale gene expression and regulatory data from human brains across several major psychiatric disorders and stages of brain development.
Through the consortium's publication of multiple studies, it seeks to shed light on poorly understood psychiatric conditions, from autism to bipolar disorder, and ultimately jumpstart new treatment approaches.
"Our study contributes to this growing body of knowledge, showing the utility of using human cells, organoids, functional screening methods, and deep learning to investigate regulatory elements and variants involved in human brain development," says Chengyu Deng, Ph.D., a postdoctoral researcher at UCSF and a co-first author of the study.
More information: Chengyu Deng et al, Massively parallel characterization of regulatory elements in the developing human cortex, Science (2024). DOI: 10.1126/science.adh0559