Researchers design variant of main painkiller receptor
Opioids, such as morphine, are still the most effective class of painkillers, but they come with unwanted side effects and can also be addictive and deadly at high doses. Designing new pain-killing drugs of this type involves testing them on their corresponding receptors, but access to meaningful quantities of these receptors that can work in experimental conditions has always been a limiting factor.
Now, an interdisciplinary collaboration between researchers at the University of Pennsylvania has developed a variant of the mu opioid receptor that has several advantages when it comes to experimentation. This variant can be grown in large quantities in bacteria and is also water-soluble, enabling experiments and applications that had previously been very challenging or impossible.
The study was led by Renyu Liu, an assistant professor in the Department of Anesthesiology and Critical Care at Penn's Perelman School of Medicine, and Jeffery Saven, an associate professor in the Department of Chemistry in the School of Arts and Sciences. Jose Manuel Perez-Aguilar, then a graduate student in the Department of Chemistry, and Jin Xi, Felipe Matsunaga and Xu Cui, lab members in the Department of Anesthesiology and Critical Care, along with Bernard Selling of Impact Biologicals Inc., contributed significantly to this study.
Their research was published in the Journal PLOS ONE.
The mu opioid receptor belongs to a class of cellular membrane proteins called G protein-coupled receptors, or GPCRs. Involved in wide range of biological processes, these receptors bind to molecules in the environment, initiating cellular signaling pathways. In the case of this receptor, binding to opioid molecules leads to a profound reduction of pain but also to a variety of unpleasant and potentially fatal side-effects, a problem that researchers from multiple disciplines are attempting to address.
"There are two directions for solving this problem in basic science, either working on the opioid molecule or working on the receptor," Liu said. "We're doing the latter."
Experimenting on the mu opioid receptor has been challenging for several reasons. The human receptor itself is relatively scarce, appearing in small quantities on only a few types of cells, making harvesting appreciable amounts impractical. Researchers have also been unable to grow it recombinantly—genetically engineering bacteria to express the protein en masse—as some parts of the protein are toxic to E. coli. Hydrophobic, or water-hating, amino acid groups on the exterior of the receptor that help it sit in the cell's membrane also make it insoluble in water when isolated.
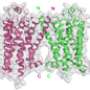
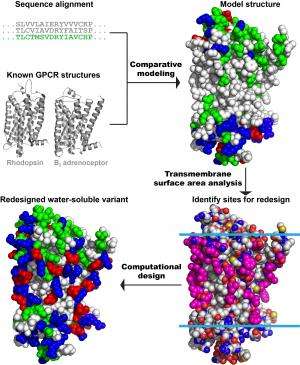
The researchers set out to address these challenges by computationally designing variants of the mu opioid receptor. This task had challenges of its own; their research was conducted long before the crystal structure of receptor was known.
"The problem with this receptor is that the native structure has only very recently been solved and only a significant re-engineered mouse model at that," Liu said. "When we started this project, we were blind."
Starting with only the gene sequence for the human version of the receptor, the researchers knew the order of the protein's amino acids but not how they were folded together. The structures for other GPCRs, such as rhodopsin and the beta-2 adrenergic receptor, were known at the time, however.
"Based on the comparison of our sequence to the sequences of those GPCRs, we built a computer model of the protein," Saven said. "When the structure of the mouse version of this receptor appeared, we were able to compare our model to that structure, and they matched up really well."
From that comparison, the researchers were able to identify the hydrophobic amino acids on the exterior of the structure, as well as some of those that were potentially toxic to E. coli.
"The objective then was to redesign those exterior amino acids," Saven said. "Based on the physical and chemical interactions these amino acids have with each other and with water, we were able to identify sequence combinations that are consistent with the model—where atoms don't overlap in space—and preferentially occupy the exterior surface with ones that are water soluble."
Replacing 53 of the protein's 288 amino acids, the research team introduced the new gene sequence into E. coli, which were able to produce large quantities of the variant. Beyond looking like the now-available mouse mu opioid receptor, the researchers were able to show its value to future studies by performing functional tests.
"We showed that this water-soluble form of the protein can compete with the native, membrane-based form when binding with antagonists that are fluorescently labeled," Saven said. "You can watch the fluorescence shift as more of these water-soluble variants are floating around in the solution."
The team's computational approach enables further iterations of the variant to be more easily designed, meaning it can be tweaked alongside experimental conditions.
"This is a great product that can do a lot of things," Liu said. "You can use this variant to look at the structure-function relationship for the receptor, or even potentially use it as a screening tool."