Genome study identifies three possible drug candidates for autoimmune diseases
(Medical Xpress)—New pharmaceuticals to fight autoimmune diseases, such as multiple sclerosis, rheumatoid arthritis and psoriasis, may be identified more effectively by adding genome analysis to standard drug screening, according to a new study by a research team led by UC San Francisco and Harvard researchers, in collaboration with Tempero and GlaxoSmithKlein.
In a study reported online April 17, 2014 in the journal Immunity, the scientists combined drug screening with state-of-the-art techniques for analyzing the genome, leading to three small molecules that improved symptoms in a mouse form of multiple sclerosis.
The three potential drug candidates, selected from a large library of screened chemicals, each knocked down the response of Th17 cells, a type of immune cell that drives many autoimmune diseases by attacking normal cells in the body. More specifically, the drugs homed in on an essential molecule within the Th17 cells.
"We examined what makes Th17 cells – which play a crucial role in multiple autoimmune diseases – distinct from other closely related T cells within the immune system," said Alexander Marson, MD, PhD, a leading T cell expert and member of the UCSF Diabetes Center. "Then we investigated several small molecules that inhibit the development and function of these cells. When the Th17 cells were hit by these molecules we saw less severe multiple-sclerosis-like symptoms in the mice."
The research team, led by Marson and Vijay K. Kuchroo, PhD, an immunologist at Brigham and Women's Hospital in Boston and Harvard Medical School, combined powerful techniques to shed light on a class of protein molecules within cells known as transcription factors.
Drug designers have rarely targeted transcription factors. Each transcription factor binds to DNA at a unique set of locations along the 23 pairs of chromosomes, and thereby influences which genes are turned on and off to trigger the protein production that drives cell development and function.
Different transcription factors shape the development of different types of T cells within the immune system, Marson and others are discovering. In their new study, Marson found that the transcription factor called ROR gamma t has a unique role in guiding development of Th17 cells, while inhibiting the development of other immune cells.
Preventing Th17 cells from developing by inhibiting the function of ROR gamma t appears to be an effective strategy for fighting autoimmune diseases, Marson said.
"There already are drugs in clinical trials for autoimmune diseases – including psoriasis and rheumatoid arthritis – that are antibodies for IL-17 or IL-17 receptors," Marson said, referring to signaling molecules secreted by Th17 cells that can help trigger an attack our own healthy tissue, and the receptors that receive those signals. "This is an entirely different and promising approach to fight autoimmune disease," he said.
"Our studies map a path to targeting transcription factors and provide both insight into how transcriptional regulators shape the identity and affect the development of Th17 cells, and also into how different drug molecules might affect these regulatory circuits in the cells," he said.
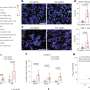
To reveal the distinct and sometimes subtle effects of the drug candidates, the researchers studied the entire genome to see where ROR gamma t attached to DNA, which genes were activated or turned off as a result, and how these effects were altered by the drug candidates.
"Not only did we look at which genes are turned on and off, but we also systematically looked at DNA-binding sites across this genome," Marson said. "This pushes the boundary of what's typically done."
In addition to attaching to DNA, ROR gamma t has a pocket that looks like it should bind a hormone, Marson said. But what this hormone might be, and its effects, are unknown. The different drug candidates that inhibited Th17 development had different effects on ROR gamma t and resultant DNA binding and gene activation, possibly because of distinct interactions with the hormone-binding pocket, Marson said.
Analyzing the large data sets generated through such experiments could help pharmaceutical companies wading into development of drugs that target transcription factors to test the waters, Marson said, enabling drug developers to better understand mechanisms of drug action and to more easily see gene activity that could trigger side effects.
According to Marson, "This is a new, broadly applicable approach for systematically evaluating leading drug candidates for autoimmune diseases."
More information: Paper: www.sciencedirect.com/science/ … ii/S1074761314001186