Gut microbes browse along a gene buffet
In the moist, dark microbial rainforest of the intestine, hundreds of species of microorganisms interact with each other and with the cells of the host animal to get the resources they need to survive and thrive.
Though there's a lot of competition in this vibrant ecosystem, collaboration is valued too. A new study on the crosstalk between microbes and cells lining the gut of mice shows just how cooperative this environment can be.
One of the main ways that hosts manage their interactions with microbes is by carefully controlling the genes that their cells use. Duke University researchers, with colleagues from UNC-Chapel Hill and Stanford, found that the host genes in the intestine are poised to respond to microbes, and the microbes signal to the host to determine which genes respond. The study appears August 7 in the journal Genome Research.
"The intestine has a tough assignment – it has to allow for digestion and absorption of dietary nutrients while also carefully harboring and managing the teeming microbial community within," said John Rawls, an associate professor of molecular genetics and microbiology in the Duke School of Medicine.
"These physiologic responsibilities and microbial interactions vary at different places along the gut." Molecules produced on demand by the host's genes have lots of different jobs that might help or hurt the bugs: immune responses, digestive enzymes, physiological "climate control" and metabolism, among others. In some cases, the microbes might even be calling in immune system attacks on their competitors, Rawls said. "Good fences make good neighbors."
Scientists have known for some time that different genes of the host are active (or expressed) at different stretches along the length of the gut, which is about 25 feet in humans. But how those genes interact with the microbial community hasn't been clear.
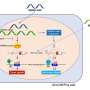
Every cell contains the complete set of DNA in the human genome, but most of it is tightly spooled away in storage and unavailable for expression. Depending on the tissue type and what jobs that tissue is doing, distinct portions of DNA are unspooled to become available for activity through a structure called open chromatin, said Gregory Crawford, an associate professor of pediatrics and expert on gene expression. These open chromatin regions are known to be key locations in the genome that control which genes are expressed and which aren't.
The researchers went into this study expecting to find that the microbes signaled the host to open up areas of chromatin to activate gene expression in the gut. But what they found is that the host chooses which chromatin regions are opened to make genes available for use in each region of the intestine.
Three different mouse populations were used for the study: mice that were raised germ-free, mice that started germ-free for 8-10 weeks but then got an intensive two-week colonization with microbes, and conventionally-raised mice exposed to whatever microbes were available in their environment.
Rather than finding three different patterns of open chromatin however, the experiments found that all three were pretty much the same. Their microbial exposures varied and gene expression patterns varied, but the parts of the genome that were open at each location in the gut remained consistent, Crawford said.
"In other words, access to the genes is determined by the host, but usage of particular genes is regulated by the microbes," Crawford said.
The current study just looked at cells of the epithelium, the layer of cells lining the gut, but there would be other cells responding to these microbial signals as well. "It's likely that these microbial signals reach other cells throughout the body, which in turn mount their own specific responses," Rawls said.
Rawls said many genes that are activated by microbes in the mouse gut are similarly responsive in the fish gut. "These are presumably very ancient modes of communication between the microbes and their animal hosts," Rawls said. "Microbes shape our health profoundly, but we're only starting to understand how they do it."
Having established some understanding of the open chromatin landscape in healthy mice, the researchers now hope to figure out how these relationships change with disease states.
More information: "Microbiota modulate transcription in the intestinal epithelium without remodeling the accessible chromatin landscape," J. Gray Camp, Christopher Frank, Colin Lickwar, Harendra Guturu, Tomas Rube, Aaron Wenger, Jenny Chen, Gill Bejerano, Gregory Crawford, John Rawls. Genome Research, Sept. 2014. DOI: 10.1101/gr.165845.113