Scientists uncover mechanism that propels liver development after birth

Any expectant mother will tell you that she wants her baby's organs to develop properly in the womb.
What she may not realize, however, is that a child's internal organs continue to develop for months and years after birth. This critical period is full of cellular changes that transform the organization and function of most tissues. But the exact mechanisms underlying postnatal organ maturation are still a mystery.
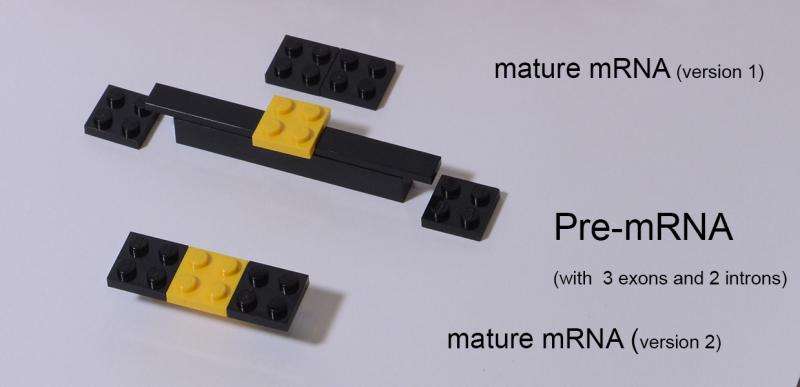
Now researchers report that liver cells utilize a mechanism called "alternative splicing," which alters how genes are translated into the proteins that guide this critical period of development.
"This mechanism is different from simply turning gene expression on or of," said University of Illinois Biochemistry Professor Auinash Kalsotra, who led the study.
"Turning gene expression on or off leads to a quantitative change in gene expression - you make more or less of a particular RNA. Alternative splicing, however, provides means to produce a qualitative change. You are making the same amounts of RNA but of different kinds."
Alternative splicing is a lot like building with LEGOs, where bits and pieces of DNA (called exons) can be pieced together or have parts removed to produce different assortments of proteins.
"The diversity of RNAs and proteins generated in this way allows the liver to acquire new functions tailored for the adult needs," Kalsotra said.
Using a powerful technology called next-generation RNA sequencing, the researchers simultaneously looked at thousands of genes, pinpointing the ones that undergo regulated changes in alternative splicing as the liver develops.
The findings, which appear in the journal Nature Communications, also identified an RNA binding protein, ESRP2, which controls this developmental program in the liver.
"It turns out that ESRP2 is absent in the fetal liver and is only turned on after birth to activate splicing of genes that are particularly important for liver growth and functionality," Kalsotra said.
Working in cell culture, the researchers spurred liver cells to express the ESRP2 protein, and observed that the cells began to display adult-like characteristics.
"We were amazed to see how clean the results were," Kalsotra said. "In the absence of ESRP2, the adult liver remains immature. This tells us how important this RNA binding protein is for optimizing adult functions."
This is the first study to provide a direct link between splicing regulation and liver maturation, he said.
"We are excited to investigate this link further and determine the exact function of these splicing switches in postnatal liver development," said Kalsotra, who also is an affiliate of the Carl R. Woese Institute for Genomic Biology at Illinois.
More information: Amruta Bhate et al. ESRP2 controls an adult splicing programme in hepatocytes to support postnatal liver maturation, Nature Communications (2015). DOI: 10.1038/NCOMMS9768