How tiny changes help T cells to survive
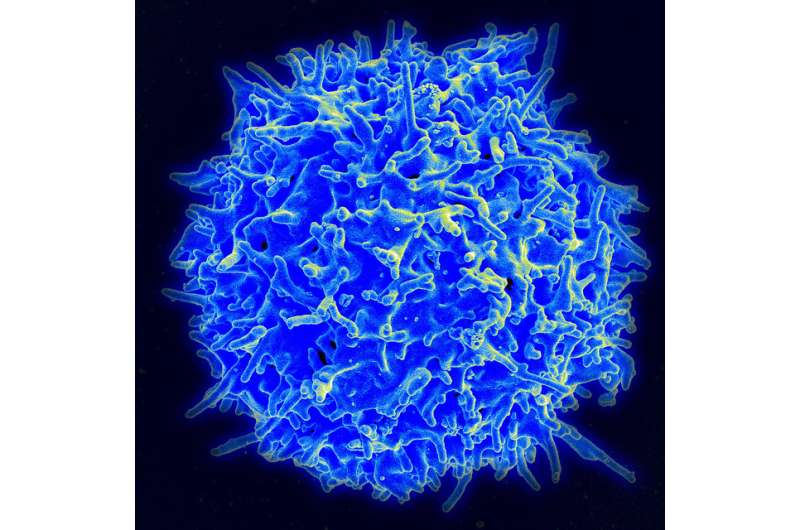
The research group led by immunologists Vigo Heissmeyer and Taku Ito-Kureha of LMU and Helmholtz Munich has revealed the essential function of m6A modifications in T cells.
The chemical modification of nucleic acids, methylation, exists not only on DNA but also on RNA. Whether this methylation is important for certain cell types and what effects it has on the interaction of cells in the body is still unclear according to current knowledge. The most abundant mRNA modification is N6-methyladenosine (also called m6A). It appears to play a crucial role in cell differentiation and to influence a number of biological processes. In m6A, a methyl group is attached to the sixth carbon atom of the nucleic base adenosine. Researchers found that a so-called writer complex is responsible for this—as in DNA. It decides which adenosine in which mRNA is modified and thus carries new information in addition to the genetic code. This information is then "read" by so-called reader proteins, which decide on the lifetime or translation of the mRNA into the encoded protein.
New success for basic research
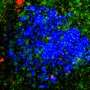
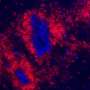
Vigo Heissmeyer and his research team investigated how the loss of m6A affects the biology of T cells. To this end, the scientists knocked out a component of the m6A writer complex in mice—both in regulatory T cells and naïve T cells. From former studies, the researchers deduced that m6A modifications are essential for T cells when they recognize an antigen. Normally, antigen recognition would lead to T cell activation, cell division and acquisition of effector functions. Regulatory T cells without m6A would lose control over the immune response. Naïve cells, on the other hand, would be arrested in their differentiation and fall into a kind of coma—that was the state of research.
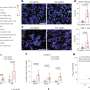
But the new study shows something different for naïve T cells: Genetic inactivation of m6A gene regulation changes the signal transduction of T cells and increases their calcium flux, making them hyperactive and inducing cell death.
The lack of m6A regulation in T cells shows a severe inflammatory response in mice. They suffer from colitis, the chronic inflammation of the intestine. This disease is triggered by an excess of activated conventional T cells. An imbalance emerges because the regulatory T cells can no longer dampen activation of the naïve ones. In the absence of m6A in T cells, a complex deregulation of the immune response occurs, naïve T cells that are activated by antigen recognition, divide initially, but also die increasingly—and can still cause the disease, presumably due to lack of suppression by regulatory T cells.
In a next step, Heissmeyer's team wants to study the molecules that recognize methylation. "Our idea is to identify new key molecules of T cell activation through insights into this type of gene regulation," Vigo Heissmeyer explains. "We can see from our study which mRNAs are modified and thus need to be precisely defined in quantity for immune responses to be error-free. If we fully understand how this regulation prevents cell death or hyperactivation, we will also know which nodes can be therapeutically manipulated."
More information: Taku Ito-Kureha et al, The function of Wtap in N6-adenosine methylation of mRNAs controls T cell receptor signaling and survival of T cells, Nature Immunology (2022). DOI: 10.1038/s41590-022-01268-1