This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
AI analysis of cancer mutations may improve therapy
Cancer has many faces—no wonder, then, that the range of cancer-causing mutations is huge as well. The totality of such genomic alterations in an individual is what experts call a "mutational landscape." These landscapes differ from one another depending on the type of cancer. And even people suffering from the same cancer often have different mutation patterns.
Researchers have already cataloged the mutational landscapes of numerous types of cancer. Somatic structural variants (SVs) have been shown to account for more than half of all cancer-driving mutations. These are those mutations in cells that emerge over the course of life—such as when copying errors creep into the DNA during cell division—and thereby alter the chromosome structure.
They are not inherited and are found only in affected cells and in their daughter cells. As we age, such genomic alterations become more numerous, and a person's mutational landscape increasingly comes to resemble a unique mosaic.
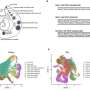
Although somatic SVs play a crucial role in cancer development, relatively little is known about them. "There is a lack of methods that analyze their effects on cell function," explains Dr. Ashley Sanders, who heads the Genome Stability and Somatic Mosaicism Lab at the Max Delbrück Center. That's changing thanks to new research findings, which Sanders recently published in the journal Nature Biotechnology along with the European Molecular Biology Laboratory (EMBL).
"We developed a computational analysis method to detect and identify the functional effects of somatic SVs," she reports. This enabled the team to understand the molecular consequences of individual somatic mutations in different leukemia patients, giving them new insights into the mutation-specific alterations. Sanders says it may also be possible to use these findings to develop therapies that target the mutated cells, adding that "they open up exciting new avenues for personalized medicine."
Even more detailed than conventional single-cell analyses
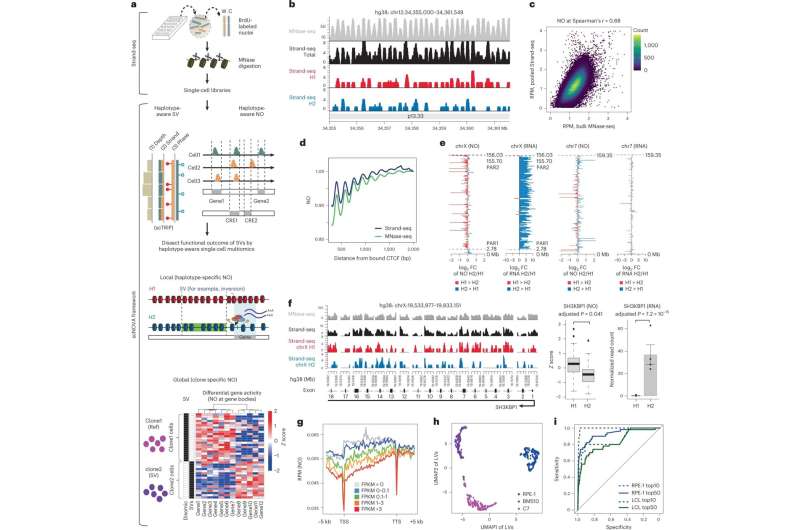
Their calculations are based on data from Strand-seq—a special single-cell sequencing method that Sanders played an instrumental role in developing and that was first introduced to the scientific community in 2012. This technique can examine a cell's genome in much greater detail than conventional single-cell sequencing technologies.
Thanks to a sophisticated experimental protocol, the Strand-seq method can independently analyze the two parental DNA strands (one from the father and one from the mother). With conventional sequencing methods, distinguishing such homologs—chromosomes that are similar in shape and structure but not identical—is nearly impossible.
"By resolving the individual homologs within a cell, somatic SVs can be identified much better than with other methods," explains Sanders. The approach used for doing this was described by the researcher and her colleagues in a paper that appeared in Nature Biotechnology in 2020.
The research team is part of the joint research focus "Single-Cell Approaches for Personalized Medicine" of the Berlin Institute of Health at Charité (BIH), Charité—Universitätsmedizin Berlin, and the Max Delbrück Center.
Building on this work, they are now able to also determine the positions of nucleosomes in each cell. Nucleosomes are units of DNA wrapped around protein complexes called histones, and play a crucial role in organizing chromosomes. The position of nucleosomes can change during gene expression, with the type of wrapping revealing whether or not a gene is active. Sanders and her colleagues developed a self-learning algorithm to compare the gene activity of patient cells with and without somatic SV mutations, allowing them to determine the molecular impact of the structural variants.
New targets for cancer therapy
"We can now take a sample from a patient, look for the mutations that led to the disease, and also learn the signaling pathways that the disease-causing mutations disrupt," explains Sanders. For example, the team was able to identify a rare but very aggressive mutation in a leukemia patient. The nucleosome analysis provided the researchers with information about the signaling pathways involved, which they used to specifically inhibit the growth of cells containing the mutation.
"This means that a single test tells us something about the cellular mechanisms involved in cancer formation," says Sanders. "We can eventually use this knowledge to develop personalized treatments, guided by each patient's unique condition."
More information: Hyobin Jeong et al, Functional analysis of structural variants in single cells using Strand-seq, Nature Biotechnology (2022). DOI: 10.1038/s41587-022-01551-4
Ester Falconer et al, DNA template strand sequencing of single-cells maps genomic rearrangements at high resolution, Nature Methods (2012). DOI: 10.1038/nmeth.2206
Ashley D. Sanders et al, Single-cell analysis of structural variations and complex rearrangements with tri-channel processing, Nature Biotechnology (2019). DOI: 10.1038/s41587-019-0366-x