This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Transcription factors found to contribute to subtypes of colorectal cancers
New research in colorectal cancers directed by investigators at the Johns Hopkins Kimmel Cancer Center suggests that expression of transcription factors—proteins that help turn specific genes on or off by binding to nearby DNA—may play a central role in the degree of DNA methylation across the genome, contributing to the development of different subtypes of these cancers. Methylation is a process in which certain chemical groups attach to areas of DNA that guide genes' on/off switches.
Studying the expression of these transcription factors in patients with colorectal cancers could reveal biomarkers to help determine overall survival in people with a subgroup of colorectal cancers who generally have better survival rates and, importantly, respond better to immune checkpoint therapy—a type of immunotherapy that releases restraints that cancer cells place on the immune response—and other treatments.
Similar patterns of transcription factor expression could be seen by the researchers even in precancerous polyps, and could potentially be used by physicians to determine which patients need closer follow-up to prevent cancer development.
A description of the work was published in the Proceedings of the National Academy of Sciences.
Aberrant DNA methylation is a well-known phenomenon occurring in cancers, explains senior study author Hariharan Easwaran, Ph.D., M.Sc., an associate professor of oncology at the Johns Hopkins Kimmel Cancer Center, but the degree of DNA methylation varies in cancers of the same tissue type.
Some colon and other cancers have a very high degree of DNA methylation gains while others have much lower frequency of DNA methylation gains, he says. Traditionally, these have been described in an area of the genome known as a promoter region, which helps launch the transcription process. The exact mechanisms underlying these changes have not been clear.
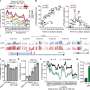
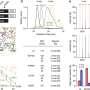
In a series of laboratory studies of genetic material taken from tubular adenomas (precancerous polyps in the colon) and colon tumors, the researchers linked cancer-specific transcription factor expression alterations to methylation alterations in colorectal cancers and their premalignant precursor lesions, which provided insights into the origins and evolution of different molecular subtypes of colorectal cancers.
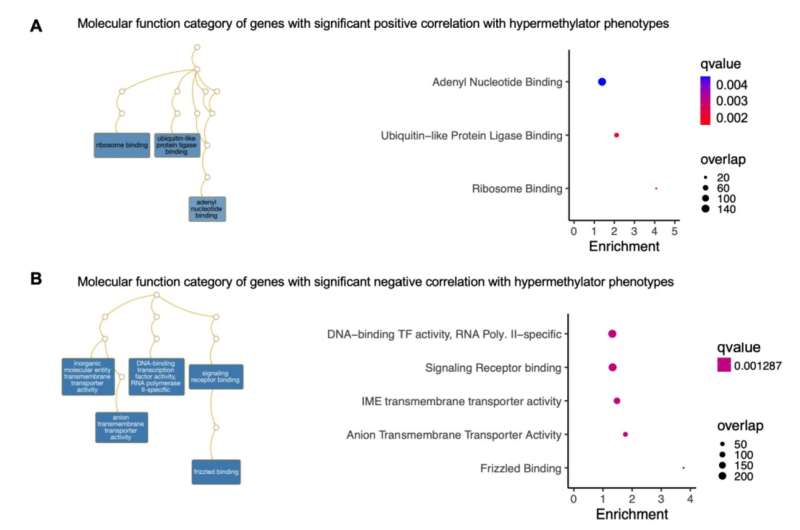
Specifically, researchers observed that some regions of the genome undergoing increased methylation tend to have binding sites for transcription factors that are downregulated, or have low expression. In some types of colon cancer, based on the types of genetic alterations associated with the cancer, transcription factors are upregulated or have higher expression.
The findings suggest that cancer-specific methylation differences potentially evolve due to perturbation in the activity or expression of transcription factors. Similar changes in DNA methylation patterns were observed in precancerous polyps.
"These studies highlight that the transcription factor expression changes and corresponding DNA methylation changes are early events during tumor development," says lead study author Yuba Bhandari, Ph.D., a research associate at the Johns Hopkins Kimmel Cancer Center.
"As polyps do not carry all of the key genetic changes typically found in full-blown cancer cells, the transcription factor changes may represent the earliest molecular regulators of precancerous cells, with profound impact on the genome-wide DNA methylation changes."
The specific set of transcription factors identified in the study may help in stratifying colorectal cancer prognosis, Easwaran adds.
"This is particularly important, because multiple studies have shown that a certain subtype of colorectal cancers responds best to immune checkpoint blockade therapies, while others may not fare as well," he says. "Expression profiling of relevant transcription factors may help develop better therapeutic strategies across subtypes of colorectal cancers."
More information: Yuba R. Bhandari et al, Transcription factor expression repertoire basis for epigenetic and transcriptional subtypes of colorectal cancers, Proceedings of the National Academy of Sciences (2023). DOI: 10.1073/pnas.2301536120