This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Study delves deep into cancer tissue to uncover an immune evasion mechanism
Smart users of social media protect themselves by deleting suspicious "friend requests" that might serve as a gateway to harmful content. Similarly, our immune system uses its own judgment to identify cellular "profiles" that seem dangerous. As immune cells patrol the body, they check out protein pieces called antigens; generated when proteins are sent to degradation, these pieces are presented on the surface of cells. When an antigen pattern looks suspicious, the immune system "hits delete" and eradicates the cell.
In cancer, regulatory processes are disrupted, increasing the likelihood that abnormal proteins will be produced, and consequently, presented as antigens on the cellular surface. Yet, despite their suspicious antigen profile, cancer cells manage to evade the immune system's watchful gaze.
A study conducted by Prof. Yifat Merbl's team in the Systems Immunology Department at the Weizmann Institute of Science has delved deep into the cellular source of antigens—the waste-processing machinery known as the proteasome—and pulled out a previously unknown mechanism that allows cancer cells to slip by immune system defenses. The findings were recently published in Nature Cancer.
Cancer: A different path to protein degradation
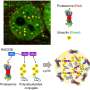
The proteasome's job is to break down worn-out or damaged proteins into shorter protein chains called peptides. Some peptides are then recycled, while others get processed into antigens, those that make up the cellular profile that is presented to the immune system.
This is not the first time Merbl has turned the spotlight on this cellular waste processing. Over the past few years, her team has developed technologies for tracking down proteins that undergo degradation by the proteasome. The resulting data is sufficiently detailed to reveal the unique structure of each degraded peptide.
In the new study, led by Aaron Javitt and Merav Shmueli, the team applied these technologies to create the first-ever map of proteasome degradation activity in patient-derived tumor cells.
"We applied our system to uncover what happens in the course of waste processing in tissues removed from patients with a common type of lung cancer," Merbl says. "When we compared peptides derived from cancer cells with those from adjacent, noncancerous tissue, we noticed differences not only in the subset of proteins that were degraded, but also in the way they had been processed and cut."
This prompted the team to examine the proteasome from a new perspective: Rather than looking at the peptides it generates, they focused on the proteasome machinery itself. "We looked at the cancerous tissue and wondered—what is different about the structure of its proteasomes?" Merbl asks.
A specialized 'cookbook' for cancer survival
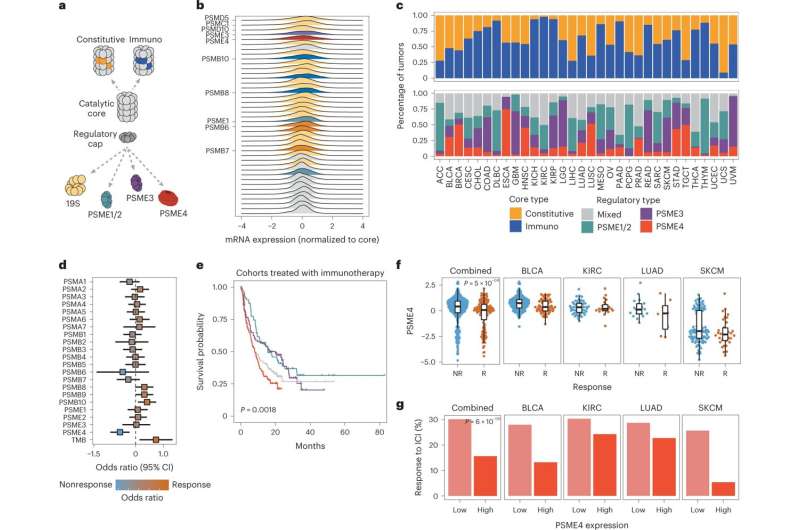
The proteasome is a protein complex organized into an empty barrel-shaped tube made up of specialized protein-degrading enzymes. This core can interact with additional cap-like subunits that govern which proteins can access the barrel for processing and the pace at which they are degraded. Different types of proteasomes are mainly classified based on the specific enzymes within the barrel, and they specialize in different types of degradation. For example, the immunoproteasome excels at producing peptides ideally suited to becoming antigens.
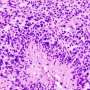
When the researchers started rummaging through the waste-processing machinery in tissues removed from patients with lung cancer, they noted an astonishingly large number of proteasomes that contained the protein PSME4. This protein, known as one of the regulatory "caps" making up the proteasome, was rarely found in proteasomes from adjacent, noncancerous tissues.
The team then set out to characterize the unique degradation style of the PSME4-enriched proteasomes. To illustrate what they found, imagine different proteasomes as chefs with different seasoning preferences. Some might prefer a sour flavor, while the immunoproteasome devises "sweeter" peptides that are particularly attractive to immune cells.
Using advanced biochemical techniques, the team discovered that higher levels of PSME4 lead to increased production of the sour peptides in a cell and a lower amount of sweet-flavored peptides. In a series of tests, the team showed that this imbalanced ratio between sweet and sour peptides hinders the ability of the immune system to accurately identify cancer, resulting in a compromised immune response.
Based on this observation, the team hypothesized that high levels of PSME4 in a tumor could weaken patient response to immunotherapy, a treatment aimed at enabling the immune system to better combat cancer.
To test their theory, the team used online databases containing information about different cancers and patients' responses to various treatments. In their search for a "proteasome fingerprint" characteristic of cancer cells, they were first of all surprised to discover just how notoriously heterogeneous the proteasome subunits in different types of cancer were. In addition, although the PSME4 protein was not enriched in all cancers, the databases clearly showed that patients whose cancers had high levels of PSME4 were less responsive to immunotherapy.
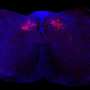
To make sure that PSME4 levels indeed directly affect the immune system, the researchers conducted a series of experiments involving mouse models of lung cancer. When mice were injected with cancer cells with reduced PSME4 expression, their immune systems were able to eliminate any signs of tumors. In contrast, injections of cancer cells with excessive PSME4 resulted in gigantic tumors and a negligible immune response. Finally, mice lacking any adaptive immune system were not affected by either an increase or a decrease in PSME4, which supported the idea that PSME4 levels affect the cancer by influencing the immune response.
"Our study focused on the proteasome in lung cancer, but our data indicate that there are other cancer types where PSME4 is abnormally abundant," says Merbl. She stresses the importance of studying the cellular waste-processing machinery, which might conceal additional mechanisms governing the interactions between cancer cells and the immune system. Her lab is seeking to develop a treatment that would reduce PSME4 levels in cancer or block its binding to the proteasome, hopefully making tumors more susceptible to immunotherapy.
While the development of such a treatment is still a future prospect, the study showed that characterizing the proteasomes of a cancerous tumor may one day assist physicians in selecting a personalized approach, that is, making treatment choices that will be appropriate for each individual patient.
More information: Aaron Javitt et al, The proteasome regulator PSME4 modulates proteasome activity and antigen diversity to abrogate antitumor immunity in NSCLC, Nature Cancer (2023). DOI: 10.1038/s43018-023-00557-4