This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Scientists identify T-cell differentiation nodes to improve cancer-killing
T cells are the key immune system component involved in killing cancer. Tumors produce signals that turn off these T cells, partially by forcing them to progressively differentiate (mature) into a hypofunctional state known as exhaustion.
Scientists from St. Jude Children's Research Hospital comprehensively examined the transcription factors involved in T-cell differentiation states in cancer. They then used this information to enhance anticancer activity in preclinical models by promoting or blocking T-cell differentiation. The findings, which have implications for cancer immunotherapy, were published today in Nature.
T cells are used in adoptive cell therapy (ACT) to target and kill cancer cells. Among ACTs, chimeric antigen receptor (CAR) T cells have shown clinical efficacy in blood cancers but have not been as effective in solid tumors. This variance in efficacy is partly because tumors promote T-cell exhaustion, in which the cells are less effective at actively killing cancer. The scientists showed they could precisely interrupt the flow of the differentiation process, thereby enhancing antitumor efficacy.
"T cells are the cornerstone of tumor immunotherapy, and we found a new way to reprogram T cells to make them more effective," said Hongbo Chi, Ph.D., St. Jude Department of Immunology. "We can push them towards a specialized state so that they can be more functional tumor-killing cells."
Modifying differentiation to flow toward cancer-killing T cells
T-cell differentiation is conceptually similar to a river running down a mountain. The top of the mountain is the precursor cell, and the bottom an exhausted cell. The desired state is between these two extremes—like having a lake in the middle of the mountain—that is differentiated enough, but not too much, to have high proliferation and effective anticancer activity. The lake receives water from above but also has a stream constantly draining it further down the slope. Uninterrupted, all water in the lake will eventually drain through the river leading down below—all the T cells will become exhausted.
The scientists found a way to increase the water flow from above and dam the river draining the lake, resulting in more water accumulation in the lake or, more aptly, more T cells that effectively kill tumors. For example, when the researchers deleted the transcription factor ETS proto-oncogene 1 (ETS1), the amount of cancer-killing T cells was increased by enhancing differentiation to the intermediate state (increasing flow into the lake).
On the other hand, knocking out the transcription factor recombination signal binding protein for immunoglobulin kappa J region (RBPJ) increased the accumulation of intermediate state T cells by blocking terminal differentiation (damming the river at the lake). Each approach showed markedly increased antitumor activity.
Even more promising, when the researchers combined their modified T cells with a type of immunotherapy called immune checkpoint blockade, they further increased antitumor activity.
"We've provided new potential strategies to enhance immunotherapy," said co-first author Peipei Zhou, Ph.D., St. Jude Department of Immunology. "We increased the amount of cancer-killing T cells by differentiating them from upstream precursors, and we could also block terminal differentiation to increase accumulation of the functional cells. Both approaches improved antitumor efficacy in our immune checkpoint blockade models."
While the work was done in mouse models, the researchers used bioinformatics data from human clinical samples to verify that these same genes are important in these processes in humans.
Mapping the differentiation journey of exhausted T cells
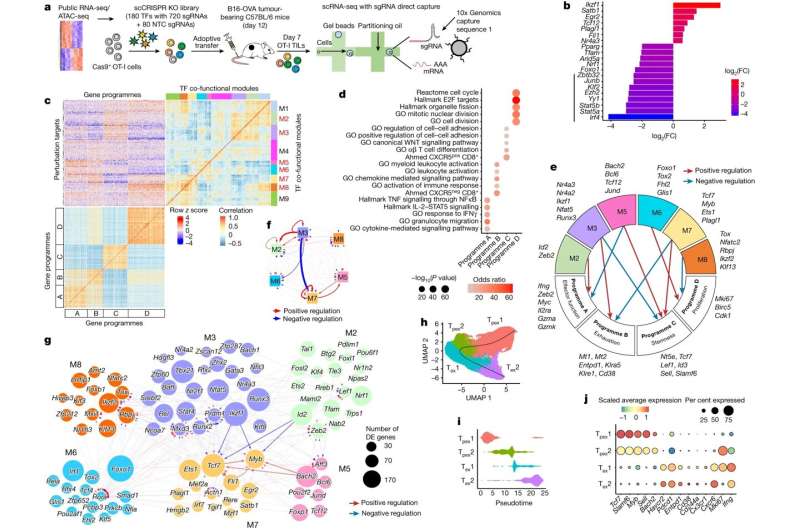
T cells infiltrate tumors to go about their work of destroying them. The researchers mapped the transcription factors expressed in the T cells infiltrating tumors. Their work showed which individual transcription factors and their networks are responsible for each T cell differentiation state, providing targets for improving immunotherapy.
"We created a map for the field of T-cell differentiation and exhaustion in tumors based on the loss-of-function genetic screening at high dimension," said co-first author Hao Shi, Ph.D., St. Jude Department of Immunology. "This map will provide a guide for future researchers to refer to and use to identify ways to improve T cell-based immunotherapies."
Where past attempts to understand T-cell exhaustion focused on an individual molecule or transcription factor, this screen was more comprehensive. This allowed the researchers to document the entirety of the gene regulatory networks involved.
"We've provided the most complete picture of the transcription factors in T cells inside a tumor ever created," Zhou said.
"Such a causal transcriptional network provides new insights into lineage differentiation, a fundamental process in biology," added Shi.
The group performed this feat using single-cell CRISPR-Cas9 screening, a gene editing technology that analyzes gene expression profiling of individual cells after selectively removing transcription factors in a comprehensive screen. Transcription factors are proteins that directly regulate gene expression. By examining an individual T cell's gene expression pattern, the researchers could compare them to see which knocked-out transcription factors most affected T-cell differentiation and anticancer activity.
"The same approach may be more broadly applicable to increase our knowledge in many biological contexts beyond T cells and immunology," Chi said. "We showed this new technology, single-cell CRISPR screening in vivo, can drive novel biological discovery. I think we're at the advent of something truly exciting."
More information: Hongbo Chi, Single-cell CRISPR screens in vivo map T cell fate regulomes in cancer, Nature (2023). DOI: 10.1038/s41586-023-06733-x. www.nature.com/articles/s41586-023-06733-x