This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Immunoengineering researchers decode the 'cytokine storm' in sepsis
Sepsis—when an infection causes the immune system to improperly target the body—is one of the leading causes of death in the ICU.
While many studies have examined the dynamics that lead to sepsis, the molecules of the immune system that are thought to produce significant damage to the body—cytokines—have not been fully understood. These proteins help control inflammation, but when the immune system responds more aggressively than it should, it can release a "cytokine storm" on all tissues, causing tissue injury, organ failure, and death.
To better understand sepsis and the role of cytokines, University of Chicago Pritzker School of Molecular Engineering (PME) researchers have measured gene expression across tissues and organs affected by sepsis in a mouse model. They then measured how those same tissues were affected by pairs of cytokines. This work was led by Michihiro Takahama, a former postdoctoral fellow in the Chevrier lab, who is now an assistant professor at Osaka University in Japan.
Surprisingly, they found that three cytokine pairs were responsible for most of the body's damaging response to sepsis.
"We created the first organism-wide map of the effect of sepsis which uncovered a hierarchy within the cytokine storm," said Asst. Prof. Nicolas Chevrier, co-author of the research. "And despite the chaotic nature of the storm, the rule that can explain this chaos turned out to be much simpler than we thought."
The results, which could ultimately lead to new therapies for the condition, are published in Nature Immunology.
Measuring gene expression across tissues
Though sepsis can be treated with antibiotics, it can lead to septic shock and death. One in three people who dies in a hospital has had sepsis, according to the Centers for Disease Control.
For Chevrier, who aims to uncover the rules that govern the immune system's behavior across the whole body, tackling the complicated factors of a dysregulated immune response like sepsis is key to understanding the system as a whole.
"We know the symptoms of sepsis, but most of the information we have is from blood draws," he said. "The knowledge that was lacking the most was what happened inside tissue at the cellular and molecular levels."
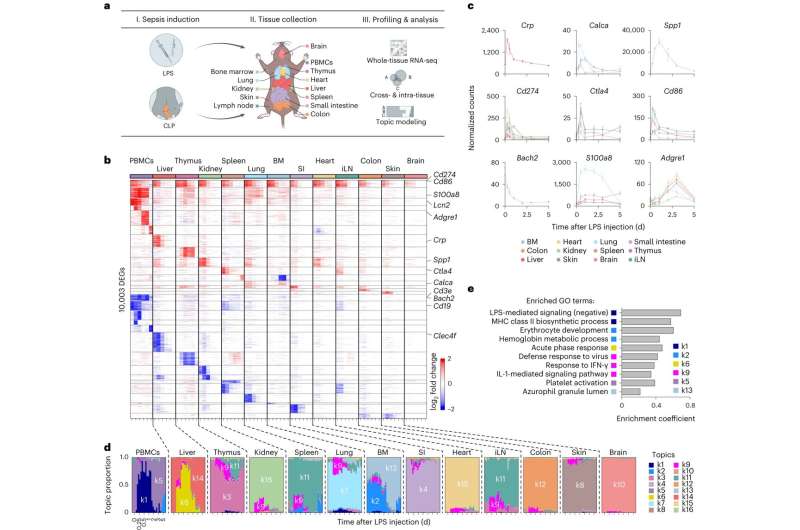
To better understand the body-wide response to sepsis, Chevrier and his team measured gene expressions across tissues in mouse models with sepsis. Measurements were taken across organs—including the brain, heart, and skin—across six points in time, to better understand the dynamic nature of the condition. The team identified more than 10,000 genes that were expressed across the body, encompassing nearly half of all the genes present in the mouse genome.
The team then wondered how much of this gene expression could be attributable to cytokines. High cytokine levels in sepsis patients are known to be associated with poor outcomes, but researchers didn't know which cells and pathways were impacted in each tissue. "It's hard to disentangle the effects of cytokines, which can have many activities depending on which mixtures of cytokines are present in any given tissue and cellular context," Chevrier said.
To try to understand which cytokines were responsible for tissue damage, the team focused on six cytokines known to play a role in sepsis, looking at them one by one.
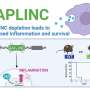
Curiously, they found that no effect from single cytokines could explain the organism-wide effects seen in sepsis. But when they began studying them in pairwise combinations, they found that three cytokines (IL-18, IFN-γ, IL-1β) paired with the cytokine TNF showed the same effect on gene expression across tissues as sepsis.
Testing to see if principle holds true in humans
The team used their whole-tissue gene expression—PME-seq—and spatial transcriptomic analyses to map the effects of these cytokine pairs on nearly 200 cell types across the body, showing the vast effect of sepsis on tissues.
In doing so, they found that non-lymphoid tissues returned to normal faster than lymphoid tissues, which might help explain why patients who survive sepsis still face lower survival rates in the following years due to impaired immune system functioning.
Next, Chevrier will test whether this cytokine pair principle holds true in human tissue, as well. If so, it could lead to new therapy avenues for sepsis. Though scientists have designed cytokine blockers to treat conditions like sepsis, they have been mostly unsuccessful. Perhaps, Chevrier says, using combinations of blockers for these specific cytokines could be key to success.
More information: Michihiro Takahama et al, A pairwise cytokine code explains the organism-wide response to sepsis, Nature Immunology (2024). DOI: 10.1038/s41590-023-01722-8