Polymerases pause to help mediate the flow of genetic information
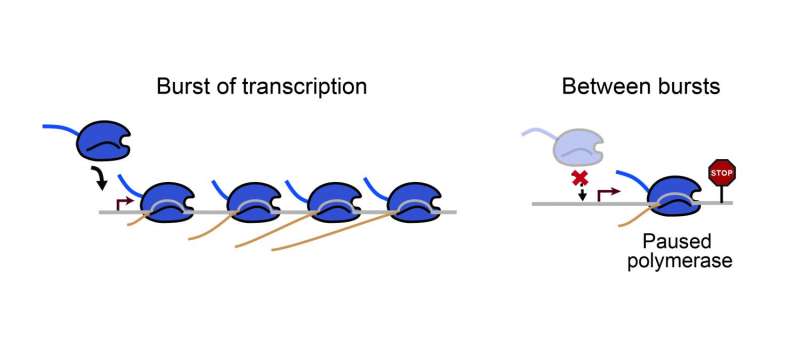
Stop-and-go traffic is typically a source of frustration, an unneccesary hold-up on the path from point A to point B. But when it comes to the molecular machinery that copies our DNA into RNA, a stop right at the beginning of the path may actually be helpful. Recent research from the Stowers Institute for Medical Research shows that this stop prevents another machine from immediately following the first, presumably to better control the traffic and avoid later collisions.
Each time a gene is "turned on" or expressed, a molecule called RNA polymerase must position itself at a specific spot along the DNA and travel down its winding strands, transcribing the gene from start to finish. Scientists once thought that as soon as a polymerase got the signal to go, it would zoom to the finish line, like a car on a race track. But then they discovered that most polymerases pause shortly after they start, without a clear reason why.
In a study published online May 15, 2017, in Nature Genetics, Stowers researchers show that when one polymerase pauses on the track, it keeps other polymerases from entering the starting gate. Paradoxically, polymerases that pause for longer periods of time can mediate faster and more synchronized gene expression in response to the kinds of signals triggered by various stages of development or dysregulated in cancer.
"We discovered a traffic rule that appears to guide the process of transcription," says Stowers Associate Investigator Julia Zeitlinger, Ph.D., who led the study. "Genes are transcribed through bursts of activity, like rush hour. Traffic is pretty dangerous. It makes sense to tightly control the number of cars on the road and minimize the number of accidents."
Genetic information flows from DNA to RNA to protein. Because transcription is the first step "to making practically everything" in the cell, Zeitlinger says, it has been an intense area of study for decades. Thousands of studies have focused on the initiation of transcription, when the polymerase first assembles itself on the DNA. But over the last ten years, scientists have come to realize that polymerases spend much of their time a short distance down from the starting gate, as if delayed by a caution flag.
Zeitlinger wondered how this pausing related to initiation, and what kind of an effect it had on the overall transcription process. In this study, Predoctoral Researcher Wanqing Shao used a method called ChIP-nexus developed by the Zeitlinger Lab to map the position of polymerases on the DNA, both in the presence and absence of drugs that block transcription. She found that paused polymerases were far more stable than polymerases assembled at the initiation site. In addition, Shao showed that paused polymerases kept new polymerases from initiating transcription.
This finding could be particularly important in the context of transcription bursts, a rapid succession of numerous transcribing polymerases interspersed by periods of inactivity that can last minutes or even hours. Since paused polymerases were observed to be so stable, the researchers think that they not only block other polymerases from immediately following them during bursts of transcription, but that they also sit there in between bursts of transcription.
"They are preventing other polymerases from lining up when the traffic lights are red and traffic is idle. The busy traffic only resumes when the cell needs more of this gene and turns the traffic lights green again," said Zeitlinger. "Having traffic rules makes sense. Leaving traffic to randomness would be inefficient and dangerous."
For example, cancer can arise when gene expression is allowed to unfold unchecked. Therefore, by understanding the basic mechanisms that control gene expression, researchers can gain a greater appreciation of the underlying causes of cancer and related diseases.
More information: Wanqing Shao et al. Paused RNA polymerase II inhibits new transcriptional initiation, Nature Genetics (2017). DOI: 10.1038/ng.3867