New characterization of human genome mutability catalyzes biomedical research
As biomedical researchers continue to make progress toward the realization of personalized genomic medicine, their focus is increasingly tuned to highly mutable regions of the human genome that contribute significantly to genetic variation and many inherited disorders.
Accurately characterizing mutability has posed a serious challenge, but a team of Penn State researchers recently took an important step toward providing a comprehensive geographic characterization of mutability in the human genome.
The results of an interdisciplinary study led by Huck Institutes of the Life Sciences affiliates Kateryna Makova and Francesca Chiaromonte will be published this week in the journal Proceedings of the National Academy of Sciences.
Other key contributors to the study were Penn State doctoral students Prabhani Kuruppumullage Don, currently a candidate in the statistics program, and Guruprasad Ananda, a graduate of the Huck Institutes' bioinformatics and genomics program who has recently accepted a position with Jackson Laboratory in Bar Harbor, Maine.
"In this project we combined genome-wide data on human-orangutan DNA differences, genetic variability within Homo sapiens, several features of the human genomic landscape, and detailed functional annotations of the human genome," said Makova, professor of biology at and director of the Center for Medical Genomics.
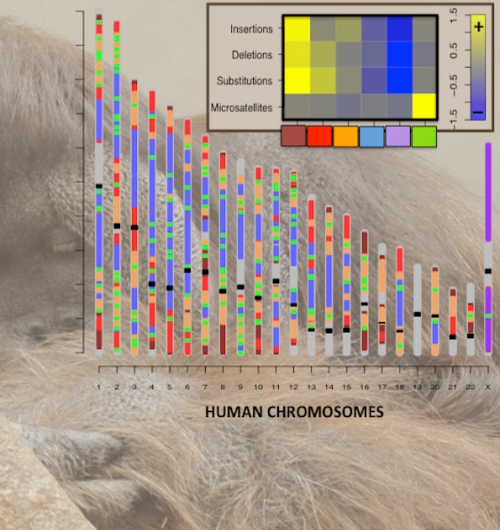
Such rich information allowed the researchers to discern regions of the genome with particular mutational regimes. For example, they found some regions where rates of different mutation types are all elevated (hot regions), and others where the rates are all reduced (cold regions).
"The location of these regions in the genome is not random and can be associated with intragenomic differences in GC content, recombination rates, methylation, etc.," said Makova. "Intriguingly, we found that protein-coding genes preferentially inhabit mutationally hot regions, likely because mutations of these genes can confer an adaptive advantage."
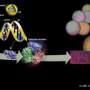
Estimating the rates of four common mutation types—nucleotide substitutions, small (? 30bp) insertions and small deletions, and mononucleotide microsatellite repeat number alterations—across the human genome, the researchers analyzed and mapped the incidence of those mutations onto corresponding chromosomal segments, yielding a genome-wide profile of mutagenetic mechanisms and potential.
"Hidden Markov Models, which have a long history of applications in genomics, were instrumental in unveiling the biological implications of our rich data," said Chiaromonte, professor of statistics and public health sciences.
Using these models, the researchers were able to quantitatively characterize the different mutational regimes, or "hidden states," and to partition the genome into contiguous segments governed by each such regime.
"Importantly, with this approach we are demarcating switches in mutational regimes along the genome—the boundaries between segments—based on the data," said Chiaromonte. "Since we utilize four mutation rates simultaneously, our results account for and exploit interdependencies among different types of change that affect the genome. We also employed simulations to assess associations between mutational regimes, genomic landscape features, and the spatial organization of functional elements."
The paper not only represents a significant contribution to scientists' understanding of the intricacies of human mutagenesis, but also provides a foundation for biomedical analyses, such as screening genomes for cancer- and other disease-related variants, which may assist in the validation of disease-causing sites across the genome and catalyze development of targeted, site-specific therapeutic strategies.
The results have far-reaching implications for several areas of biomedical sciences, according to Makova.
"First, knowledge about mutationally hot and cold regions can aid in screening disease variants, since hot regions are expected to give more false positives," she said. "Second, previous studies demonstrated that mutation rates are usually overestimated when pedigree data are used; we show that such overestimation occurs because of mutations located in hot regions. Third, information about mutationally hot and cold regions can improve predictions of functional noncoding elements in the genome, which are expected to be less conserved in mutationally hot regions. Ultimately, we and other researchers can utilize the results of our analysis, which are publicly available, to address these pressing questions in medical, evolutionary and functional genomics."