How a thieving transcription factor dominates the genome
One powerful DNA-binding protein, the transcription factor PU.1, steals away other transcription factors and recruits them for its own purposes, effectively dominating gene regulation in developing immune cells, according to a new Caltech-led study. The research was conducted in the laboratory of Ellen Rothenberg, Albert Billings Ruddock Professor of Biology. A paper describing the work appears in the June 19 issue of the journal Immunity.
At some point in a stem cell's life, it must make an irreversible transition to develop into a particular cell type. When this happens, its nucleus begins to bustle with activity: Proteins called transcription factors latch onto the cell's DNA at specific locations (called binding sites) to tune up and down the expression of various genes. This act of gene regulation is the key to the diversity of cells we see in the human body—neurons, muscle cells, immune cells. However, how transcription factors determine whether they will turn genes on or off is still poorly understood.
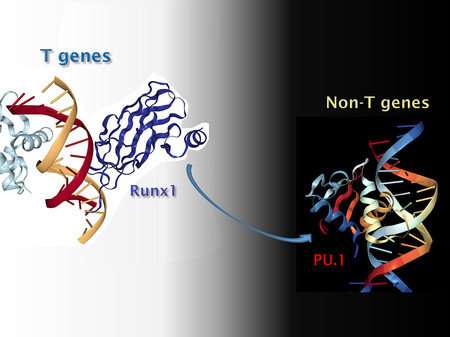
Now, a particular transcription factor called PU.1 has been found to steal transcription factors away from their other jobs, not only forcing them to work together with PU.1 but also effectively turning off the genes that those factors were previously working upon.
"Our finding helps us understand how stem cells make decisions about differentiation," says Hiroyuki Hosakawa, senior postdoctoral scholar and the study's co-first author. "Though we specifically studied T cells, we expect that the behavior of these transcription factors is broadly similar to any kind of cell, like muscle or motor neurons. PU.1 seems to be a kind of master transcription factor influencing gene expression."
Genes are like blueprints, encoding for the proteins that build a cell and give it functionality. The first step in the process of decoding a gene is called transcription, where a given gene is read and copied into an RNA form for use in protein synthesis. If a cell needs a lot of a particular protein, its corresponding gene will be upregulated. If a cell no longer needs a protein, that corresponding gene will be downregulated or turned off.
Transcription factors, as their name suggests, regulate transcription by latching onto binding sites on DNA. There are hundreds of different kinds of transcription factors, each factor able to contribute to the control of multiple genes, and each type of factor recognizing a different DNA sequence, called a binding site. These sites are locations where factors can bind with other factors to work together in combinations and modify gene expression. It has been theorized that transcription factors only work on the genes located next to a factor's binding site. However, sometimes the genes that a factor can physically bind to are not the same genes that actually change their expression in response to that factor. This can happen, according to the new research, because of the way transcription factors compete with one another.
One particular transcription factor, PU.1, is only present in early stages of the development of stem cells into T cells. It can have a powerful influence: The whole pattern of gene expression in the cells changes when PU.1 disappears from the cells, which is a key milestone in T cell development. Thus, PU.1 is a versatile regulator, able to either crank up gene transcription or suppress it. Strangely though, at the actual sites where it binds, PU.1 only seems to turn genes on; how it keeps other genes off has been a mystery. Rothenberg and her team aimed to determine how this happens.
The researchers discovered that PU.1 steals other transcription factors. When PU.1 is not present, other transcription factors go about their business, binding to their respective sites on the genome. When PU.1 enters the scene, several other types of transcription factors are drawn to it, neglecting their functions on other genes. As a result, those genes turn off while the genes PU.1 regulates are boosted. Thus, during the early developmental stages when it is present, PU.1 is able to dial down expression of genes that it does not even physically bind to.
"We found that transcription factors can actually control gene expression without directly binding to genes," says co-first author and former postdoctoral scholar Jonas Ungerbäck, now of Lund University in Sweden.
"It explains a lot of mysteries about the fine tuning of biological developmental states," says Rothenberg. "Biological systems are very sensitive, so as soon as there is competition—like between transcription factors for binding—you can have ripple effects. For PU.1 to actually affect the activity of another factor that's being removed from its normal function, it shows that this factor is very powerful in an unexpected way."
The paper is titled "Transcription factor PU.1 represses and activates gene expression in early T cells by redirecting partner transcription factor binding."
More information: Hiroyuki Hosokawa et al. Transcription Factor PU.1 Represses and Activates Gene Expression in Early T Cells by Redirecting Partner Transcription Factor Binding, Immunity (2018). DOI: 10.1016/j.immuni.2018.04.024